Structural insights into GrpEL1-mediated nucleotide and substrate release of human mitochondrial Hsp70
- PMID: 39737924
- PMCID: PMC11685456
- DOI: 10.1038/s41467-024-54499-1
Structural insights into GrpEL1-mediated nucleotide and substrate release of human mitochondrial Hsp70
Abstract
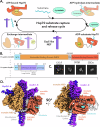
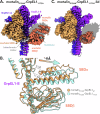
Maintenance of protein homeostasis is necessary for cell viability and depends on a complex network of chaperones and co-chaperones, including the heat-shock protein 70 (Hsp70) system. In human mitochondria, mitochondrial Hsp70 (mortalin) and the nucleotide exchange factor (GrpEL1) work synergistically to stabilize proteins, assemble protein complexes, and facilitate protein import. However, our understanding of the molecular mechanisms guiding these processes is hampered by limited structural information. To elucidate these mechanistic details, we used cryoEM to determine structures of full-length human mortalin-GrpEL1 complexes in previously unobserved states. Our structures and molecular dynamics simulations allow us to delineate specific roles for mortalin-GrpEL1 interfaces and to identify steps in GrpEL1-mediated nucleotide and substrate release by mortalin. Subsequent analyses reveal conserved mechanisms across bacteria and mammals and facilitate a complete understanding of sequential nucleotide and substrate release for the Hsp70 chaperone system.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Inclusion & Ethics statement: The research described here includes local researchers from the University of California, San Diego. The roles and responsibilities of this research were agreed upon by all included authors.
Figures






Update of
-
Structural insights into GrpEL1-mediated nucleotide and substrate release of human mitochondrial Hsp70.bioRxiv [Preprint]. 2024 May 13:2024.05.10.593630. doi: 10.1101/2024.05.10.593630. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10815. doi: 10.1038/s41467-024-54499-1. PMID: 38798347 Free PMC article. Updated. Preprint.
References
-
- Kim, Y. E., Hipp, M. S., Bracher, A., Hayer-Hartl, M. & Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem.82, 323–355 (2013). - PubMed
-
- Voos, W. Chaperone–protease networks in mitochondrial protein homeostasis. Biochim. Biophys. Acta BBA Mol. Cell Res.1833, 388–399 (2013). - PubMed
-
- Morán Luengo, T., Mayer, M. P. & Rüdiger, S. G. D. The Hsp70–Hsp90 chaperone cascade in protein folding. Trends Cell Biol.29, 164–177 (2019). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R35 GM138206/GM/NIGMS NIH HHS/United States
- T32-GM008326/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35-GM138206/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 GM129325/GM/NIGMS NIH HHS/United States
- T32 GM008326/GM/NIGMS NIH HHS/United States

