Immune checkpoint inhibitor-induced severe epidermal necrolysis mediated by macrophage-derived CXCL10 and abated by TNF blockade
- PMID: 39737932
- PMCID: PMC11685864
- DOI: 10.1038/s41467-024-54180-7
Immune checkpoint inhibitor-induced severe epidermal necrolysis mediated by macrophage-derived CXCL10 and abated by TNF blockade
Abstract
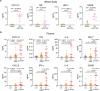
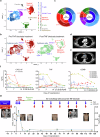
Immune checkpoint inhibitors (ICI) represent new anticancer agents and have been used worldwide. However, ICI can potentially induce life-threatening severe cutaneous adverse reaction (SCAR), such as Stevens-Johnson syndrome/toxic epidermal necrolysis (SJS/TEN), hindering continuous ICI therapy. We examine 6 cohorts including 25 ICI-induced SJS/TEN patients and conduct single-cell RNA sequencing (scRNA-seq) analysis, which shows overexpression of macrophage-derived CXCL10 that recruits CXCR3+ cytotoxic T lymphocytes (CTL) in blister cells from ICI-SJS/TEN skin lesions. ScRNA expression profiles and ex vivo blocking studies further identify TNF signaling as a pathway responsible for macrophage-derived CXCL10 and CTL activation. Based on the trajectory analysis, ICI-activated T cells from whole blood are proposed to serve as the initial cells involved in inflammation, that lead to monocytes differentiating into macrophages and increasing their susceptibility to migrate to the lesion sites. Compared with systemic corticosteroids treatment, ICI-induced SJS/TEN patients treated with biologic TNF blockade showed a significantly rapid recovery and no recurrence of SCAR with continuous ICI therapy. Our findings identify that macrophage-eliciting CTL contribute to the pathogenesis of ICI-induced epidermal necrolysis and provide potential therapeutic targets for the management and prevention of SCAR induced by ICI therapy.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

