ITGB4/CD104 mediates zika virus attachment and infection
- PMID: 39737945
- PMCID: PMC11685869
- DOI: 10.1038/s41467-024-54479-5
ITGB4/CD104 mediates zika virus attachment and infection
Erratum in
-
Author Correction: ITGB4/CD104 mediates zika virus attachment and infection.Nat Commun. 2025 Apr 7;16(1):3303. doi: 10.1038/s41467-025-58610-y. Nat Commun. 2025. PMID: 40195301 Free PMC article. No abstract available.
Abstract
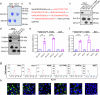
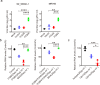
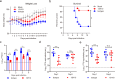
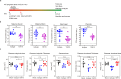
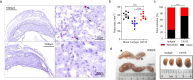
Zika virus (ZIKV) infection can result in a birth defect of the brain called microcephaly and other severe fetal brain defects. ZIKV enters the susceptible host cells by endocytosis, which is mediated by the interaction of the envelope (E) glycoprotein with cellular surface receptor molecules. However, the cellular factors that used by the ZIKV to gain access to host cells remains elusive. Here, we report that the extracellular domain of integrin beta 4 (ITGB4) is an entry factor of ZIKV. ITGB4 mediates ZIKV infection by directly interacting with the E glycoprotein of ZIKV, and ITGB4 knockout hampers the binding and replication of ZIKV to host cells. A functional monoclonal antibody against ITGB4 or the soluble forms of ITGB4 could decrease the binding and infection of ZIKV to permissive cell lines. Importantly, the ITGB4 antibody blocks the infection of ZIKV to mouse placenta, thus protecting the fetuses from ZIKV infection. Together, our study has demonstrated that ZIKV infection involves ITGB4 dependent binding.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 32060039/National Natural Science Foundation of China (National Science Foundation of China)
- 8227142882030033, 92254301, and 81921006/National Natural Science Foundation of China (National Science Foundation of China)
- 2022SHZR1769/Natural Science Foundation of Inner Mongolia (Inner Mongolian Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

