NOS inhibition sensitizes metaplastic breast cancer to PI3K inhibition and taxane therapy via c-JUN repression
- PMID: 39737957
- PMCID: PMC11685991
- DOI: 10.1038/s41467-024-54651-x
NOS inhibition sensitizes metaplastic breast cancer to PI3K inhibition and taxane therapy via c-JUN repression
Abstract
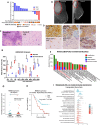
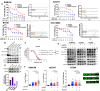
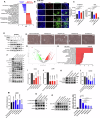
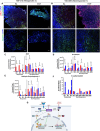
Metaplastic breast cancer (MpBC) is a highly chemoresistant subtype of breast cancer with no standardized therapy options. A clinical study in anthracycline-refractory MpBC patients suggested that nitric oxide synthase (NOS) inhibitor NG-monomethyl-l-arginine (L-NMMA) may augment anti-tumor efficacy of taxane. We report that NOS blockade potentiated response of human MpBC cell lines and tumors to phosphoinositide 3-kinase (PI3K) inhibitor alpelisib and taxane. Mechanistically, NOS blockade leads to a decrease in the S-nitrosylation of c-Jun NH2-terminal kinase (JNK)/c-Jun complex to repress its transcriptional output, leading to enhanced tumor differentiation and associated chemosensitivity. As a result, combined NOS and PI3K inhibition with taxane targets MpBC stem cells and improves survival in patient-derived xenograft models relative to single-/dual-agent therapy. Similarly, biopsies from MpBC tumors that responded to L-NMMA+taxane therapy showed a post-treatment reversal of epithelial-to-mesenchymal transition and decreased stemness. Our findings suggest that combined inhibition of iNOS and PI3K is a unique strategy to decrease chemoresistance and improve clinical outcomes in MpBC.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: C. Yam reports grants from Conquer Cancer Foundation (Career Development Award supported by Fleur Fairman; the Gianni Bonadonna Breast Cancer Research Fellowship) and other support from MD Anderson Cancer Center (The University of Texas, Houston, TX) during the conduct of the study, as well as other support from Amgen, Merck, Genentech, and GSK outside the submitted work. S. Moulder also reports other support from Eli Lilly and Company and grants from ASCO Career Development Award Mentor outside the submitted work. J. Chang reports grants from the Cancer Prevention and Research Institute of Texas and from NIH during the conduct of the study. J.Chang is the sole inventor on patent application no. 10420838 entitled “Methods for treating cancer using iNOS-inhibitory compositions” held by Houston Methodist Hospital. C. Thomas reports support from NIH during this study. F. Meric-Bernstam reports grants and personal fees from F. Hoffmann-La Roche/Genentech during the conduct of the study, as well as personal fees from AbbVie, Aduro BioTech Inc., Alkermes, AstraZeneca, DebioPharm, eFFECTOR Therapeutics, IBM Watson, Infinity Pharmaceuticals, The Jackson Laboratory, Kolon Life Science, Lengo Therapeutics, OrigiMed, PACT Pharma, Parexel International, Pfizer Inc., Samsung Bioepis, Seattle Genetics Inc., Tallac Therapeutics, Tyra Biosciences, Xencor, Zymeworks, Black Diamond, Biovica, Eisai, Immunomedics, Inflection Biosciences, Karyopharm Therapeutics, Loxo Oncology, Mersana Therapeutics, OnCusp Therapeutics, Puma Biotechnology Inc., Silverback Therapeutics, Spectrum Pharmaceuticals, and Zentalis and grants from Aileron Therapeutics, Inc., AstraZeneca, Bayer Healthcare Pharmaceutical, Calithera Biosciences Inc., Curis Inc., CytomX Therapeutics Inc., Daiichi Sankyo Co. Ltd., Debiopharm International, eFFECTOR Therapeutics, Guardant Health Inc., Klus Pharma, Takeda Pharmaceutical, Novartis, Puma Biotechnology Inc., and Taiho Pharmaceutical Co. outside the submitted work. No disclosures were reported by other authors.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

