A high-protein diet-responsive gut hormone regulates behavioral and metabolic optimization in Drosophila melanogaster
- PMID: 39737959
- PMCID: PMC11685984
- DOI: 10.1038/s41467-024-55050-y
A high-protein diet-responsive gut hormone regulates behavioral and metabolic optimization in Drosophila melanogaster
Abstract
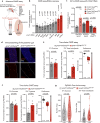
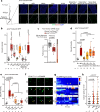
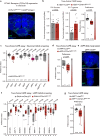
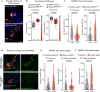
Protein is essential for all living organisms; however, excessive protein intake can have adverse effects, such as hyperammonemia. Although mechanisms responding to protein deficiency are well-studied, there is a significant gap in our understanding of how organisms adaptively suppress excessive protein intake. In the present study, utilizing the fruit fly, Drosophila melanogaster, we discover that the peptide hormone CCHamide1 (CCHa1), secreted by enteroendocrine cells in response to a high-protein diet (HPD), is vital for suppressing overconsumption of protein. Gut-derived CCHa1 is received by a small subset of enteric neurons that produce short neuropeptide F, thereby modulating protein-specific satiety. Importantly, impairment of the CCHa1-mediated gut-enteric neuronal axis results in ammonia accumulation and a shortened lifespan under HPD conditions. Collectively, our findings unravel the crosstalk of gut hormone and neuronal pathways that orchestrate physiological responses to prevent and adapt to dietary protein overload.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

