SARS-CoV-2 infection of human pluripotent stem cell-derived vascular cells reveals smooth muscle cells as key mediators of vascular pathology during infection
- PMID: 39737992
- PMCID: PMC11685814
- DOI: 10.1038/s41467-024-54917-4
SARS-CoV-2 infection of human pluripotent stem cell-derived vascular cells reveals smooth muscle cells as key mediators of vascular pathology during infection
Abstract
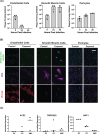
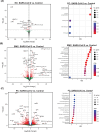
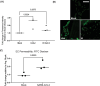
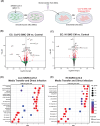
Although respiratory symptoms are the most prevalent disease manifestation of infection by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), nearly 20% of hospitalized patients are at risk for thromboembolic events. This prothrombotic state is considered a key factor in the increased risk of stroke, which is observed clinically during both acute infection and long after symptoms clear. Here, we develop a model of SARS-CoV-2 infection using human-induced pluripotent stem cell-derived endothelial cells (ECs), pericytes (PCs), and smooth muscle cells (SMCs) to recapitulate the vascular pathology associated with SARS-CoV-2 exposure. Our results demonstrate that perivascular cells, particularly SMCs, are a susceptible vascular target for SARS-CoV-2 infection. Utilizing RNA sequencing, we characterize the transcriptomic changes accompanying SARS-CoV-2 infection of SMCs, PCs, and ECs. We observe that infected SMCs shift to a pro-inflammatory state and increase the expression of key mediators of the coagulation cascade. Further, we show human ECs exposed to the secretome of infected SMCs produce hemostatic factors that contribute to vascular dysfunction despite not being susceptible to direct infection. The findings here recapitulate observations from patient sera in human COVID-19 patients and provide mechanistic insight into the unique vascular implications of SARS-CoV-2 infection at a cellular level.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests. R.J. is an advisor/co-founder of Fate Therapeutics and Fulcrum Therapeutics. D.J.M. has sponsored research, consults, and/or has stock options/stock in Medicenna, Lyell, Attivare, Epoulosis, Limax Biosciences, Lightning Bio, and Oddity Tech, licensed intellectual property with Alkem and Amend Surgical, and Board of Directors, ATCC.The remaining authors (A.R., A.K., T.W., M.F., X.G., L.G., R.K., T.L., Z.W.) declare no competing interests.
Figures

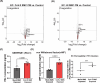
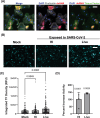
Update of
-
SARS-CoV-2 infection of human pluripotent stem cell-derived vascular cells reveals smooth muscle cells as key mediators of vascular pathology during infection.bioRxiv [Preprint]. 2024 Nov 26:2023.08.06.552160. doi: 10.1101/2023.08.06.552160. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10754. doi: 10.1038/s41467-024-54917-4. PMID: 37609322 Free PMC article. Updated. Preprint.
References
-
- WHO, Vol. 2021.
-
- Merschel, M. American Heart Association News Stories.https://www.cbsnews.com/tag/american-heart-association/ (2023).
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

