Orthogonal and multiplexable genetic perturbations with an engineered prime editor and a diverse RNA array
- PMID: 39737993
- PMCID: PMC11685949
- DOI: 10.1038/s41467-024-55134-9
Orthogonal and multiplexable genetic perturbations with an engineered prime editor and a diverse RNA array
Erratum in
-
Author Correction: Orthogonal and multiplexable genetic perturbations with an engineered prime editor and a diverse RNA array.Nat Commun. 2025 Feb 11;16(1):1535. doi: 10.1038/s41467-025-56839-1. Nat Commun. 2025. PMID: 39934174 Free PMC article. No abstract available.
Abstract
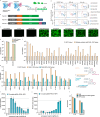
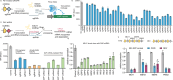
Programmable and modular systems capable of orthogonal genomic and transcriptomic perturbations are crucial for biological research and treating human genetic diseases. Here, we present the minimal versatile genetic perturbation technology (mvGPT), a flexible toolkit designed for simultaneous and orthogonal gene editing, activation, and repression in human cells. The mvGPT combines an engineered compact prime editor (PE), a fusion activator MS2-p65-HSF1 (MPH), and a drive-and-process multiplex array that produces RNAs tailored to different types of genetic perturbation. mvGPT can precisely edit human genome via PE coupled with a prime editing guide RNA and a nicking guide RNA, activate endogenous gene expression using PE with a truncated single guide RNA containing MPH-recruiting MS2 aptamers, and silence endogenous gene expression via RNA interference with a short-hairpin RNA. We showcase the versatility of mvGPT by simultaneously correcting a c.3207C>A mutation in the ATP7B gene linked to Wilson's disease, upregulating the PDX1 gene expression to potentially treat Type I diabetes, and suppressing the TTR gene to manage transthyretin amyloidosis. In addition to plasmid delivery, we successfully utilize various methods to deliver the mvGPT payload, demonstrating its potential for future in vivo applications.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: Q.C. Yuan, H.Z., T.C.D., and X.G. have submitted a provisional patent application to the U.S. Patent Office pertaining to the genetic perturbation technologies described within this work (application number 63/712,648). The remaining authors declare no competing interests.
Figures


Similar articles
-
Fusion guide RNAs for orthogonal gene manipulation with Cas9 and Cpf1.Nat Commun. 2017 Nov 23;8(1):1723. doi: 10.1038/s41467-017-01650-w. Nat Commun. 2017. PMID: 29167440 Free PMC article.
-
Guide RNAs: A Glimpse at the Sequences that Drive CRISPR-Cas Systems.Cold Spring Harb Protoc. 2016 Jul 1;2016(7). doi: 10.1101/pdb.top090902. Cold Spring Harb Protoc. 2016. PMID: 27371605
-
PrimeDesign software for rapid and simplified design of prime editing guide RNAs.Nat Commun. 2021 Feb 15;12(1):1034. doi: 10.1038/s41467-021-21337-7. Nat Commun. 2021. PMID: 33589617 Free PMC article.
-
CRISPR-Based Technologies: Impact of RNA-Targeting Systems.Mol Cell. 2018 Nov 1;72(3):404-412. doi: 10.1016/j.molcel.2018.09.018. Mol Cell. 2018. PMID: 30388409 Free PMC article. Review.
-
Improving CRISPR Genome Editing by Engineering Guide RNAs.Trends Biotechnol. 2019 Aug;37(8):870-881. doi: 10.1016/j.tibtech.2019.01.009. Epub 2019 Mar 4. Trends Biotechnol. 2019. PMID: 30846198 Review.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

