Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease
- PMID: 39737997
- PMCID: PMC11686214
- DOI: 10.1038/s41467-024-55062-8
Integrative determination of atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington disease
Abstract
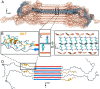
Neurodegeneration in Huntington's disease (HD) is accompanied by the aggregation of fragments of the mutant huntingtin protein, a biomarker of disease progression. A particular pathogenic role has been attributed to the aggregation-prone huntingtin exon 1 (HTTex1), generated by aberrant splicing or proteolysis, and containing the expanded polyglutamine (polyQ) segment. Unlike amyloid fibrils from Parkinson's and Alzheimer's diseases, the atomic-level structure of HTTex1 fibrils has remained unknown, limiting diagnostic and treatment efforts. We present and analyze the structure of fibrils formed by polyQ peptides and polyQ-expanded HTTex1 in vitro. Atomic-resolution perspectives are enabled by an integrative analysis and unrestrained all-atom molecular dynamics (MD) simulations incorporating experimental data from electron microscopy (EM), solid-state NMR, and other techniques. Alongside the use of prior data, we report magic angle spinning NMR studies of glutamine residues of the polyQ fibril core and surface, distinguished via hydrogen-deuterium exchange (HDX). Our study provides a molecular understanding of the structure of the core as well as surface of aggregated HTTex1, including the fuzzy coat and polyQ-water interface. The obtained data are discussed in context of their implications for understanding the detection of such aggregates (diagnostics) as well as known biological properties of the fibrils.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







Update of
-
Integrative determination of the atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington's disease.bioRxiv [Preprint]. 2024 Sep 15:2023.07.21.549993. doi: 10.1101/2023.07.21.549993. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10793. doi: 10.1038/s41467-024-55062-8. PMID: 37502911 Free PMC article. Updated. Preprint.
References
-
- Bates, G. P. et al. Huntington disease. Nat. Rev. Dis. Prim.1, 1–21 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

