The structural basis of protective and nonprotective human monoclonal antibodies targeting the parainfluenza virus type 3 hemagglutinin-neuraminidase
- PMID: 39738006
- PMCID: PMC11686389
- DOI: 10.1038/s41467-024-55101-4
The structural basis of protective and nonprotective human monoclonal antibodies targeting the parainfluenza virus type 3 hemagglutinin-neuraminidase
Abstract
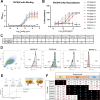
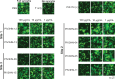
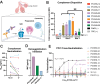
Parainfluenza virus 3 (PIV3) infection poses a substantial risk to vulnerable groups including infants, the elderly, and immunocompromised individuals, and lacks effective treatments or vaccines. This study focuses on targeting the hemagglutinin-neuraminidase (HN) protein, a structural glycoprotein of PIV3 critical for viral infection and egress. With the objective of targeting these activities of HN, we identified eight neutralizing human monoclonal antibodies (mAbs) with potent effects on viral neutralization, cell-cell fusion inhibition, and complement deposition. Three epitopes on PIV3 HN were delineated and one epitope, Site 2, elicits a mAb with cross-neutralizing ability against PIV1 and PIV3. Cryo-EM revealed the cross-neutralizing mAb utilizes a long CDR3 loop to bind inside the pocket of the sialic acid binding site. Additionally, we resolved the structure of a non-protective mAb binding to Site 1 near the HN:F-interaction site. The potent Site 2-directed mAb demonstrated clinical efficacy in hamsters, reducing viral replication prophylactically and therapeutically. These findings advance our understanding of PIV3 immunity and underscore the significance of targeting HN for clinical therapeutic development against PIV3.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: J.J.M. is an inventor on a patent application related to anti-parainfluenza virus monoclonal antibodies.
Figures



References
-
- Human Parainfluenza National Trends - NREVSS | CDC [Internet]. https://www.cdc.gov/surveillance/nrevss/human-paraflu/natl-trend.html (2023).
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- figshare/10.6084/m9.figshare.27904791
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

