Genomic analysis of global Plasmodium vivax populations reveals insights into the evolution of drug resistance
- PMID: 39738010
- PMCID: PMC11685768
- DOI: 10.1038/s41467-024-54964-x
Genomic analysis of global Plasmodium vivax populations reveals insights into the evolution of drug resistance
Abstract
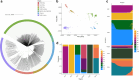
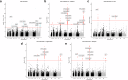
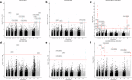
Increasing reports of chloroquine resistance (CQR) in Plasmodium vivax endemic regions have led to several countries, including Indonesia, to adopt dihydroarteminsin-piperaquine instead. However, the molecular drivers of CQR remain unclear. Using a genome-wide approach, we perform a genomic analysis of 1534 P. vivax isolates across 29 endemic countries, detailing population structure, patterns of relatedness, selection, and resistance profiling, providing insights into potential drivers of CQR. Selective sweeps in a locus proximal to pvmdr1, a putative marker for CQR, along with transcriptional regulation genes, distinguish isolates from Indonesia from those in regions where chloroquine remains highly effective. In 106 isolates from Indonesian Papua, the epicentre of CQR, we observe an increasing prevalence of novel SNPs in the candidate resistance gene pvmrp1 since the introduction of dihydroartemisinin-piperaquine. Overall, we provide novel markers for resistance surveillance, supported by evidence of regions under recent directional selection and temporal analysis in this continually evolving parasite.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- World Health Organisation. World Malaria Report 2023.
Publication types
MeSH terms
Substances
Grants and funding
- BB/T008709/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- IAA2129, MR/R026297/1, and MR/X005895/1/RCUK | Medical Research Council (MRC)
- IAA2129, MR/R026297/1, and MR/X005895/1/RCUK | Medical Research Council (MRC)
- EP/Y018842/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
- EP/Y018842/1/RCUK | Engineering and Physical Sciences Research Council (EPSRC)
LinkOut - more resources
Full Text Sources

