A new type III effector from Bradyrhizobium sp. DOA9 encoding a putative SUMO-protease blocks nodulation in Arachis hypogaea L
- PMID: 39738104
- PMCID: PMC11685577
- DOI: 10.1038/s41598-024-78913-2
A new type III effector from Bradyrhizobium sp. DOA9 encoding a putative SUMO-protease blocks nodulation in Arachis hypogaea L
Abstract
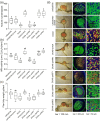
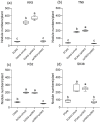
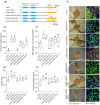
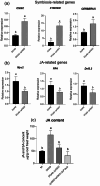
Effector proteins secreted via the type III secretion system (T3SS) of nitrogen-fixing rhizobia are key determinants of symbiotic compatibility in legumes. Previous report revealed that the T3SS of Bradyrhizobium sp. DOA9 plays negative effects on Arachis hypogaea symbiosis. In this study, we characterized the symbiotic role of 4 effector proteins (p0490, p0871, SkP48, and p0903) containing the small ubiquitin-like modifier (SUMO) protease domain identified in DOA9 during symbiosis. While the DOA9 strain and the two mutants of SUMO-proteases, p0490 and p0871, induced inefficient nodulation in A. hypogaea, the mutation of SUMO-proteases SkP48 or p0903 promoted efficient symbiosis comparable to the type strain Bradyrhizobium arachidis CCBAU051107. Complementation study of ∆p0903 with various mutated forms of p0903 highlighted importance of ubiquitin-like protein (ULP) domain in restriction of nodulation in A. hypogaea. We observed the accumulation of jasmonic acid (JA) and upregulation of several defence genes involved in the JA/ethylene (ET) signalling pathway at the early stage of infection in roots inoculated with DOA9 strain compared with those inoculated with the DOA9-∆p0903 strain. Our data highlight the importance of SUMO-protease effectors during the symbiotic interaction between bradyrhizobia and A. hypogaea, which could be useful for the development of high-performance inocula to improve its growth.
Keywords: Arachis hypogaea; Bradyrhizobium sp; T3SS; and SUMO-protease.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures


References
-
- Prakamhang, J. et al. Proposed some interactions at molecular level of PGPR coinoculated with Bradyrhizobium diazoefficiens USDA110 and B. Japonicum THA6 on soybean symbiosis and its potential of field application. Appl. Soil. Ecol.85, 38–49 (2015).
-
- Vicario, J. C., Primo, E. D., Dardanelli, M. S. & Giordano, W. Promotion of peanut growth by co-inoculation with selected strains of Bradyrhizobium and Azospirillum. J. Plant. Growth Regul.35, 413–419 (2016).
-
- Ayalew, T., Yoseph, T., Petra, H. & Cadisch, G. Yield response of field-grown cowpea varieties to Bradyrhizobium inoculation. Agron. J.113, 3258–3268 (2021).
Publication types
MeSH terms
Substances
Grants and funding
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- N11A670769/JSPS-NRCT by National Research Council of Thailand
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- B16F640113/The NSRF via the Program Management Unit for Human Resources & Institutional Development, Research and Innovation
- ANR-20-CE20-0012/French National Research Agency
- ANR-20-CE20-0012/French National Research Agency
LinkOut - more resources
Full Text Sources

