Multimodal cadmium resistance and its regulatory networking in Pseudomonas aeruginosa strain CD3
- PMID: 39738119
- PMCID: PMC11685661
- DOI: 10.1038/s41598-024-80754-y
Multimodal cadmium resistance and its regulatory networking in Pseudomonas aeruginosa strain CD3
Abstract
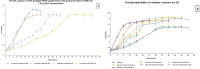
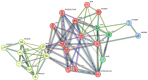
Cadmium, a toxic heavy metal, poses significant global concern. A strain of the genus Pseudomonas, CD3, demonstrating significant cadmium resistance (up to 3 mM CdCl2.H2O) was identified from a pool of 26 cadmium-resistant bacteria isolated from cadmium-contaminated soil samples from Malda, India. The minimum inhibitory concentrations (MICs) for cadmium and other heavy metals/metalloids were determined with clarity using a modified chemically-defined medium inoculated with variable inoculum density. Formation of biofilm enabled CD3 cells to resist up to 0.75 mM CdCl2.H2O. Survival and growth of CD3 cells in presence of > 1 mM CdCl2.H2O was dependent on efflux mechanism. Efflux mechanism in CD3 was confirmed by atomic absorption spectroscopy. Resistance to cadmium was inducible when grown in presence of ≥ 1.0 mM CdCl2.H2O. Minimum concentration of cadmium or zinc or cobalt salts required for induction of cadmium resistance was determined. Whole-genome-based phylogenetic tools identified CD3 as the closest relative to Pseudomonas aeruginosa DSM50071T. Bioinformatic analyses revealed a complex network of regulations, with BfmR playing a crucial role in the functions of CzcR and CzcS, essential for biofilm formation and receptor signalling pathways. Comparative genomics and mutation landscape analyses of cadmium-resistance genes in P. aeruginosa strains revealed dynamism in evolution of cadmium resistance.
Keywords: Pseudomonas aeruginosa; Biofilm; CD3; Cadmium; CzcCBA; Induction.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures








Similar articles
-
CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa.J Biol Chem. 2004 Mar 5;279(10):8761-8. doi: 10.1074/jbc.M312080200. Epub 2003 Dec 15. J Biol Chem. 2004. PMID: 14679195
-
Whole genome sequencing and comparative genomic analyses of Pseudomonas aeruginosa strain isolated from arable soil reveal novel insights into heavy metal resistance and codon biology.Curr Genet. 2022 Aug;68(3-4):481-503. doi: 10.1007/s00294-022-01245-z. Epub 2022 Jun 28. Curr Genet. 2022. PMID: 35763098
-
Characterization of cadmium-resistant bacteria and their potential for reducing accumulation of cadmium in rice grains.Sci Total Environ. 2016 Nov 1;569-570:97-104. doi: 10.1016/j.scitotenv.2016.06.121. Epub 2016 Jun 21. Sci Total Environ. 2016. PMID: 27341110
-
Isolation of toxic metal-tolerant bacteria from soil and examination of their bioaugmentation potentiality by pot studies in cadmium- and lead-contaminated soil.Int Microbiol. 2018 Jun;21(1-2):35-45. doi: 10.1007/s10123-018-0003-4. Epub 2018 Apr 11. Int Microbiol. 2018. PMID: 30810918
-
High-quality draft genome sequence of Pseudomonas aeruginosa san ai, an environmental isolate resistant to heavy metals.Extremophiles. 2019 Jul;23(4):399-405. doi: 10.1007/s00792-019-01092-w. Epub 2019 Apr 5. Extremophiles. 2019. PMID: 30949775
References
-
- Fergusson, J. The Heavy Elements: Chemistry, Environmental Impact and Health Effects (Pergamon Press, 1990).
-
- Sun, L. et al. Levels, sources, and spatial distribution of heavy metals in soils from a typical coal industrial city of Tangshan, China. Catena (Amst)175, 101–109. 10.1016/j.catena.2018.12.014 (2019).
-
- Song, H. et al. Metal distribution and biological diversity of crusts in paddy fields polluted with different levels of cadmium. Ecotoxicol. Environ. Saf.184, 109620. 10.1016/j.ecoenv.2019.109620 (2019). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

