NS1 binding protein regulates stress granule dynamics and clearance by inhibiting p62 ubiquitination
- PMID: 39738171
- PMCID: PMC11686067
- DOI: 10.1038/s41467-024-55446-w
NS1 binding protein regulates stress granule dynamics and clearance by inhibiting p62 ubiquitination
Erratum in
-
Author Correction: NS1 binding protein regulates stress granule dynamics and clearance by inhibiting p62 ubiquitination.Nat Commun. 2025 Feb 11;16(1):1531. doi: 10.1038/s41467-025-56909-4. Nat Commun. 2025. PMID: 39934148 Free PMC article. No abstract available.
Abstract
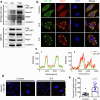
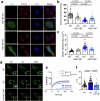
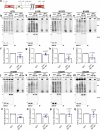
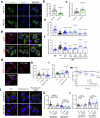
The NS1 binding protein, known for interacting with the influenza A virus protein, is involved in RNA processing, cancer, and nerve cell growth regulation. However, its role in stress response independent of viral infections remains unclear. This study investigates NS1 binding protein's function in regulating stress granules during oxidative stress through interactions with GABARAP subfamily proteins. We find that NS1 binding protein localizes to stress granules, interacting with core components, GABARAP proteins, and p62, a protein involved in autophagy. In cells lacking NS1 binding protein, stress granule dynamics are altered, and p62 ubiquitination is increased, suggesting impaired stress granule degradation. Overexpression of NS1 binding protein reduces p62 ubiquitination. In amyotrophic lateral sclerosis patient-derived neurons, reduced NS1 binding protein and p62 disrupt stress granule morphology. These findings identify NS1 binding protein as a negative regulator of p62 ubiquitination and a facilitator of GABARAP recruitment to stress granules, implicating it in stress granule regulation and amyotrophic lateral sclerosis pathogenesis.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 2020M3E5D9079911/National Research Foundation of Korea (NRF)
- 2023R1A2C2008092/National Research Foundation of Korea (NRF)
- 2023R1A2C2008092/National Research Foundation of Korea (NRF)
- RS-2023-00218515/National Research Foundation of Korea (NRF)
- JP19H05706/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Medical
Research Materials

