Rapid lightsheet fluorescence imaging of whole Drosophila brains at nanoscale resolution by potassium acrylate-based expansion microscopy
- PMID: 39738207
- PMCID: PMC11685761
- DOI: 10.1038/s41467-024-55305-8
Rapid lightsheet fluorescence imaging of whole Drosophila brains at nanoscale resolution by potassium acrylate-based expansion microscopy
Abstract
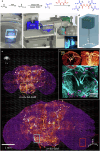
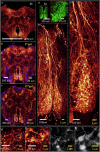
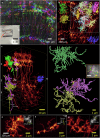
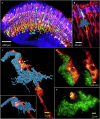
Taking advantage of the good mechanical strength of expanded Drosophila brains and to tackle their relatively large size that can complicate imaging, we apply potassium (poly)acrylate-based hydrogels for expansion microscopy (ExM), resulting in a 40x plus increased resolution of transgenic fluorescent proteins preserved by glutaraldehyde fixation in the nervous system. Large-volume ExM is realized by using an axicon-based Bessel lightsheet microscope, featuring gentle multi-color fluorophore excitation and intrinsic optical sectioning capability, enabling visualization of Tm5a neurites and L3 lamina neurons with photoreceptors in the optic lobe. We also image nanometer-sized dopaminergic neurons across the same intact iteratively expanded Drosophila brain, enabling us to measure the 3D expansion ratio. Here we show that at a tile scanning speed of ~1 min/mm3 with 1012 pixels over 14 hours, we image the centimeter-sized fly brain at an effective resolution comparable to electron microscopy, allowing us to visualize mitochondria within presynaptic compartments and Bruchpilot (Brp) scaffold proteins distributed in the central complex, enabling robust analyses of neurobiological topics.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

