An AMBRA1, ULK1 and PP2A regulatory network regulates cytotoxic T cell differentiation via TFEB activation
- PMID: 39738384
- PMCID: PMC11685475
- DOI: 10.1038/s41598-024-82957-9
An AMBRA1, ULK1 and PP2A regulatory network regulates cytotoxic T cell differentiation via TFEB activation
Abstract
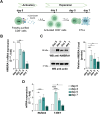
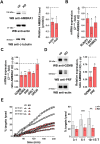
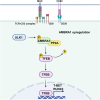
The scaffold protein AMBRA1, which participates in the autophagy pathway, also promotes CD4+ T cell differentiation to Tregs independent of autophagy through its interactor PP2A. Here we have investigated the role of AMBRA1 in CD8+ T cell differentiation to cytotoxic T cells (CTL). AMBRA1 depletion in CD8+ T cells was associated with impaired expression of the transcription factors RUNX3 and T-BET that drive CTL differentiation and resulted in impaired acquisition of cytotoxic potential. These effects were recapitulated by pharmacological inhibition of the AMBRA1 activator ULK1 or its interactor PP2A. Based on the ability of PP2A to activate TFEB, we hypothesized a role for TFEB in the CTL differentiation program regulated by AMBRA1. We show that TFEB modulates RUNX3 and T-BET expression and the generation of killing-competent CTLs, and that AMBRA1 depletion, or ULK1 or PP2A inhibition, suppresses TFEB activity. These data highlight a role for AMBRA1, ULK1 and PP2A in CTL generation, mediated by TFEB, which we identify as a new pioneering transcription factor in the CTL differentiation program.
Keywords: AMBRA1; Cytotoxic T cell; Lytic granule /; PP2A / ULK1.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- ERC_2021_SyG 951329 - ATTACK/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- IG 2017-20148/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- PRIN bando 2017-2017FS5SHL/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- PRIN 2022 Bando PNRR P2022TJZYZ/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- GR-2021-12372771/Ministero della Salute (Ministry of Health, Italy)
- PRIN bando 2017-2017FS5SHL/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- IG 2019-23543/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 0070834/Novo Nordisk Fonden (Novo Nordisk Foundation)
- R231-A14034/Dansk Kræftforskningsfond Fond (Danish Cancer Research Fund)
LinkOut - more resources
Full Text Sources
Research Materials

