Aligning knowledge concepts to whole slide images for precise histopathology image analysis
- PMID: 39738468
- PMCID: PMC11685412
- DOI: 10.1038/s41746-024-01411-2
Aligning knowledge concepts to whole slide images for precise histopathology image analysis
Abstract
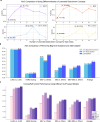
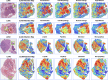
Due to the large size and lack of fine-grained annotation, Whole Slide Images (WSIs) analysis is commonly approached as a Multiple Instance Learning (MIL) problem. However, previous studies only learn from training data, posing a stark contrast to how human clinicians teach each other and reason about histopathologic entities and factors. Here, we present a novel knowledge concept-based MIL framework, named ConcepPath, to fill this gap. Specifically, ConcepPath utilizes GPT-4 to induce reliable disease-specific human expert concepts from medical literature and incorporate them with a group of purely learnable concepts to extract complementary knowledge from training data. In ConcepPath, WSIs are aligned to these linguistic knowledge concepts by utilizing the pathology vision-language model as the basic building component. In the application of lung cancer subtyping, breast cancer HER2 scoring, and gastric cancer immunotherapy-sensitive subtyping tasks, ConcepPath significantly outperformed previous SOTA methods, which lacked the guidance of human expert knowledge.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778 (IEEE, 2016).
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

