Description and phylogenetic implications of a de novo mitochondrial genome of Rhinichthys atratulus (Teleostei: Leuciscidae) from Connecticut
- PMID: 39738627
- PMCID: PMC11686256
- DOI: 10.1038/s41598-024-83134-8
Description and phylogenetic implications of a de novo mitochondrial genome of Rhinichthys atratulus (Teleostei: Leuciscidae) from Connecticut
Erratum in
-
Publisher Correction: Description and phylogenetic implications of a de novo mitochondrial genome of Rhinichthys atratulus (Teleostei: Leuciscidae) from Connecticut.Sci Rep. 2025 Feb 19;15(1):6055. doi: 10.1038/s41598-025-89662-1. Sci Rep. 2025. PMID: 39972060 Free PMC article. No abstract available.
Abstract
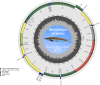
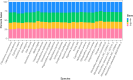
We present a de novo mitogenome assembly from a specimen of Rhinichthys atratulus, the Eastern Blacknose Dace, collected in the Connecticut River drainage. R. atratulus is a fish species widely distributed across Atlantic slope drainages from Nova Scotia, Canada to the Roanoke River Drainage, Virginia, United States. The assembly has a total length of 16,646 bp; consists of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a 974 bp D-loop; and has a GC content of 45.7%. We performed two phylogenetic analyses using the de novo assembly and 28 additional cyprinoid mitochondrial genomes: with and without the D-loop. The resulting phylogenetic trees showed the same branching patterns. However, inclusion of the D-loop resulted in nodes with equal or greater support. The trees largely match previously published analyses, while offering new insights into relationships among leuciscid genera. Our findings indicate that the inclusion of the D-loop in mitogenome phylogenetic analyses can help improve bootstrap support at otherwise unresolved nodes.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Osborne, M. J., Barela Hudgell, M. A., Caeiro-Dias, G. & Turner, T. F. The complete mitochondrial genomes of two imperiled species endemic to the Southwestern United States: Peppered Chub (Macrhybopsis tetranema) and Gila Trout (Oncorhynchus gilae). Mitochondrial DNA Part B8, 809–814. 10.1080/23802359.2023.2241658 (2023). - PMC - PubMed
-
- Nicholls, T. J. & Minczuk, M. In D-loop: 40 years of mitochondrial 7S DNA. Exp. Gerontol.56, 175–181. 10.1016/j.exger.2014.03.027 (2014). - PubMed
-
- Saccone, C. et al. Evolution of the mitochondrial genetic system: An overview. Gene261, 153–159. 10.1016/S0378-1119(00)00484-4 (2000). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

