Impact of land-use and fecal contamination on Escherichia populations in environmental samples
- PMID: 39738760
- PMCID: PMC11685573
- DOI: 10.1038/s41598-024-83594-y
Impact of land-use and fecal contamination on Escherichia populations in environmental samples
Abstract
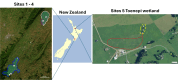
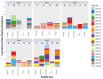
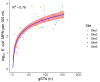
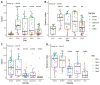
Understanding the composition of complex Escherichia coli populations from the environment is necessary for identifying strategies to reduce the impacts of fecal contamination and protect public health. Metabarcoding targeting the hypervariable gene gnd was used to reveal the complex population diversity of E. coli and phenotypically indistinct Escherichia species in water, soil, sediment, aquatic biofilm, and fecal samples from native forest and pastoral sites. The resulting amplicons were cross-referenced against a database containing over 700 different partial gnd sequences from E. coli/non-E. coli Escherichia species. Alpha and beta measures of diversity of Escherichia populations were lowest in feces, soil and sediment compared to water and aquatic biofilm samples. Escherichia populations recovered from extensive freshwater catchments dominated by sheep, beef and dairy farming were extremely diverse but well-separated from a wetland dairy site. In contrast, Escherichia populations from the low-impact native forest site with fewer fecal sources were less diverse. Metabarcoding distinguished E. coli populations important to fecal contamination monitoring from non-E. coli Escherichia environmental populations. These data represent in-depth analysis and geographic stability of Escherichia populations from environmental samples with extensive heterogeneity, and reveal links with diverse fecal sources, land-use and the overall burden of fecal contamination at sample sites.
Keywords: Fecal indicator bacteria; Freshwater; Land-use; Microbial community profiling; Source tracking; Zoonotic pathogens.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures

References
-
- Tenaillon, O., Skurnik, D., Picard, B. & Denamur, E. The population genetics of Escherichia coli. Nat. Rev. Microbiol.8, 207–217. 10.1038/nrmicro2298 (2010). - PubMed
-
- Anon. National Policy Statement for Freshwater Management 2014 (amended 2017). Ministry for the Environment. Wellington, New Zealand. https://www.mfe.govt.nz/publications/fresh-water/national-policy-stateme... (2017).
-
- Anon. National Policy Statement for Freshwater Management 2020, Ministry for the Environment. Wellington, New Zealand. https://environment.govt.nz/publications/national-policy-statement-for-f... (2020).
-
- Till, D., McBride, G., Ball, A., Taylor, K. & Pyle, E. Large-scale freshwater microbiological study: Rationale, results and risks. J. Water Health6, 443–460. 10.2166/wh.2008.071 (2008). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

