Calpain 2 promotes Lenvatinib resistance and cancer stem cell traits via both proteolysis-dependent and independent approach in hepatocellular carcinoma
- PMID: 39739077
- PMCID: PMC11688263
- DOI: 10.1186/s43556-024-00242-7
Calpain 2 promotes Lenvatinib resistance and cancer stem cell traits via both proteolysis-dependent and independent approach in hepatocellular carcinoma
Abstract
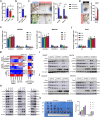
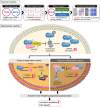
Lenvatinib, an approved first-line regimen, has been widely applied in hepatocellular carcinoma (HCC). However, clinical response towards Lenvatinib was limited, emphasizing the importance of understanding the underlying mechanism of its resistance. Herein, we employed integrated bioinformatic analysis to identify calpain-2 (CAPN2) as a novel key regulator for Lenvatinib resistance in HCC, and its expression greatly increased in both Lenvatinib-resistant HCC cell lines and clinical samples. Further in vitro and in vivo experiments indicated that knocking down CAPN2 greatly sensitized HCC cells to Lenvatinib treatment, while overexpression of CAPN2 achieved opposite effects in a Lenvatinib-sensitive HCC cell line. Interestingly, we observed a close relationship between CAPN2 expression and cancer stem cell (CSC) traits in HCC cells, evidenced by impaired sphere-forming and CSC-related marker expressions after CAPN2 knockdown, and verse vice. Mechanistically, we strikingly discovered that CAPN2 exerted its function by both enzyme-dependent and enzyme-independent manner simultaneously: activating β-Catenin signaling through its enzyme activity, and preventing GLI1/GLI2 degradation through direct binding to YWHAE in an enzyme-independent manner, which disrupting the association between YWHAE and GLI1/GLI2 to inhibit YWHAE-induced degradation of GLIs. Notably, further co-immunoprecipitation assays revealed that YWHAE could promote the protein stability of CAPN2 via recruiting a deubiquitinase COPS5 to prevent ubiquitination-induced degradation of CAPN2. In summary, our data demonstrated that CAPN2 promoted Lenvatinib resistance via both catalytic activity-dependent and -independent approaches. Reducing CAPN2 protein rather than inhibiting its activity might be a promising strategy to improve Lenvatinib treatment efficiency in HCC.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Fudan University Shanghai Cancer Center Research Ethics Committee (Approved Number: 050432–4-2108*), and all individuals provided informed consent for inclusion of their tissue in this study. Moreover, animal study was approved by the Animal Experimentation Ethics Committee of Shanghai Cancer Center, Fudan University (Approved Number: FUSCC-IACUC-S2022-0209). Competing interests: The authors declare that they have no competing interests.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
