Inhibition of CDH11 Activates cGAS-STING by Stimulating Branched Chain Amino Acid Catabolism and Mitigates Lung Metastasis of Adenoid Cystic Carcinoma
- PMID: 39739317
- PMCID: PMC11848559
- DOI: 10.1002/advs.202408751
Inhibition of CDH11 Activates cGAS-STING by Stimulating Branched Chain Amino Acid Catabolism and Mitigates Lung Metastasis of Adenoid Cystic Carcinoma
Erratum in
-
Correction to "Inhibition of CDH11 Activates cGAS-STING by Stimulating Branched Chain Amino Acid Catabolism and Mitigates Lung Metastasis of Adenoid Cystic Carcinoma".Adv Sci (Weinh). 2025 Jul;12(25):e2504992. doi: 10.1002/advs.202504992. Epub 2025 May 29. Adv Sci (Weinh). 2025. PMID: 40439487 Free PMC article. No abstract available.
Abstract
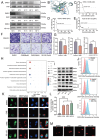
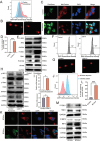
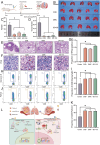
Salivary adenoid cystic carcinoma (SACC) is an intractable malignant tumor originates in the secretory glands and frequently metastasizes to the lungs. Hybrid epithelial-mesenchymal transition (EMT) cells within the tumors are correlated with augmented proliferative capacity and facilitation of lung metastasis. Single-cell RNA sequencing and spatial transcriptomic sequencing are employed to reveal the hybrid EMT subsets within the vascular fibroblast microenvironment. These hybrid EMT cells exhibit a pro-tumorigenic impact in vitro. Notably, cadherin 11 (CDH11), a specific marker for hybrid EMT cells, may exert its regulatory role in cellular function by interfering with branched-chain amino acids (BCAA) metabolism by inhibiting branched-chain ketoacid dehydrogenase to activate the mammalian target of the rapamycin pathway, thus making it a potential therapeutic target for SACC. Furthermore, celecoxib and its derivatives are specific CDH11 inhibitors that regulate BCAA metabolism, increase reactive oxygen species production, and subsequently activate the cyclic GMP-AMP synthase-stimulator of the interferongene pathway (cGAS-STING). They also inhibit lung metastasis in NOD-SCID mice in vivo. Overall, these findings suggest a promising treatment strategy that targets hybrid EMT cells to mitigate lung metastasis in SACC. Celecoxib may serve as a promising clinical intervention for the treatment of lung metastases in patients with SACC.
Keywords: adenoid cystic carcinoma; celecoxib; hybrid epithelial‐mesenchymal transforming cells; single‐cell transcriptome sequencing; spatial transcriptome sequencing; targeted therapy.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
-
- Ho A. S., Ochoa A., Jayakumaran G., Zehir A., Valero Mayor C., Tepe J., Makarov V., Dalin M. G., He J., Bailey M., Montesion M., Ross J. S., Miller V. A., Chan L., Ganly I., Dogan S., Katabi N., Tsipouras P., Ha P., Agrawal N., Solit D. B., Futreal P. A., El Naggar A. K., Reis‐Filho J. S., Weigelt B., Ho A. L., Schultz N., Chan T. A., Morris L. G., J. Clin. Invest. 2019, 129, 4276. - PMC - PubMed
-
- Bjørndal K., Krogdahl A., Therkildsen M. H., Overgaard J., Johansen J., Kristensen C. A., Homøe P., Sørensen C. H., Andersen E., Bundgaard T., Primdahl H., Lambertsen K., Andersen L. J., Godballe C., Oral Oncol. 2011, 47, 677. - PubMed
-
- Vander Poorten V. L., Balm A. J., Hilgers F. J., Tan I. B., Loftus‐Coll B. M., Keus R. B., Hart A. A., Cancer 1999, 85, 2255. - PubMed
-
- Ciccolallo L., Licitra L., Cantú G., Gatta G., Oral Oncol. 2009, 45, 669. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
