Novel Naphthyridones Targeting Pannexin 1 for Colitis Management
- PMID: 39739600
- PMCID: PMC11831487
- DOI: 10.1002/advs.202411538
Novel Naphthyridones Targeting Pannexin 1 for Colitis Management
Abstract
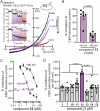
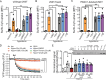
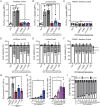
Pannexin 1 (PANX1) forms cell-surface channels capable of releasing signaling metabolites for diverse patho-physiological processes. While inhibiting dysregulated PANX1 has been proposed as a therapeutic strategy for many pathological conditions, including inflammatory bowel disease (IBD), low efficacy, or poor specificity of classical PANX1 inhibitors introduces uncertainty for their applications in basic and translational research. Here, hit-to-lead optimization is performed and a naphthyridone, compound 12, is identified as a new PANX1 inhibitor with an IC50 of 0.73 µm that does not affect pannexin-homologous LRRC8/SWELL1 channels. Using structure-activity relationship analysis, mutagenesis, cell thermal shift assays, and molecular docking, it is revealed that compound 12 directly engages PANX1 Trp74 residue. Using a dextran sodium sulfate mouse model of IBD, it is found that compound 12 markedly reduced colitis severity, highlighting new PANX1 inhibitors as a proof-of-concept treatment for IBD. These data describe the mechanism of action for a new PANX1 inhibitor, uncover the binding site for future drug design, and present a targeted strategy for treating IBD.
Keywords: LRRC8; Pannexin; colitis; inflammatory bowel disease; naphthyridone; structure–activity relationship; trovafloxacin.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Activation of neuronal P2X7 receptor-pannexin-1 mediates death of enteric neurons during colitis.Nat Med. 2012 Mar 18;18(4):600-4. doi: 10.1038/nm.2679. Nat Med. 2012. PMID: 22426419 Free PMC article.
-
Unexpected link between an antibiotic, pannexin channels and apoptosis.Nature. 2014 Mar 20;507(7492):329-34. doi: 10.1038/nature13147. Epub 2014 Mar 12. Nature. 2014. PMID: 24646995 Free PMC article.
-
Intrinsic properties and regulation of Pannexin 1 channel.Channels (Austin). 2014;8(2):103-9. doi: 10.4161/chan.27545. Epub 2014 Jan 13. Channels (Austin). 2014. PMID: 24419036 Free PMC article. Review.
-
Pannexin 1 channels facilitate communication between T cells to restrict the severity of airway inflammation.Immunity. 2021 Aug 10;54(8):1715-1727.e7. doi: 10.1016/j.immuni.2021.06.014. Epub 2021 Jul 21. Immunity. 2021. PMID: 34283971 Free PMC article.
-
[Distribution and regulation of Panx1 protein].Sheng Li Ke Xue Jin Zhan. 2013 Jun;44(3):188-92. Sheng Li Ke Xue Jin Zhan. 2013. PMID: 24027825 Review. Chinese.
References
-
- Di Virgilio F., Dal Ben D., Sarti A. C., Giuliani A. L., Falzoni S., Immunity 2017, 47, 15. - PubMed
-
- Verkhratsky A., Burnstock G., BioEssays 2014, 36, 697. - PubMed
-
- Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W., J. Cell Sci. 2007, 120, 3772. - PubMed
-
- a) Medina C. B., Mehrotra P., Arandjelovic S., Perry J. S. A., Guo Y., Morioka S., Barron B., Walk S. F., Ghesquiere B., Krupnick A. S., Lorenz U., Ravichandran K. S., Nature 2020, 580, 130; - PMC - PubMed
- b) Narahari A. K., Kreutzberger A. J., Gaete P. S., Chiu Y. H., Leonhardt S. A., Medina C. B., Jin X., Oleniacz P. W., Kiessling V., Barrett P. Q., Ravichandran K. S., Yeager M., Contreras J. E., Tamm L. K., Bayliss D. A., Elife 2021, 10, 64787. - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 113-2628-B-007-002-MY3 to Y.-H.C and H.-P.H/National Science and Technology Council
- 113-2113-M-400-001- to H.-P.H/National Science and Technology Council
- 113-2314-B-002-237-MY2 to M.-T.W. and Y.-H.C/National Science and Technology Council
- 113-BIH044 to M.-T.W/National Taiwan University Hospital
- BP-113-SP-07 to J.-C.L/National Health Research Institutes
LinkOut - more resources
Full Text Sources
