TRIM21 Promotes Tumor Growth and Gemcitabine Resistance in Pancreatic Cancer by Inhibiting EPHX1-Mediated Arachidonic Acid Metabolism
- PMID: 39739616
- PMCID: PMC11848624
- DOI: 10.1002/advs.202413674
TRIM21 Promotes Tumor Growth and Gemcitabine Resistance in Pancreatic Cancer by Inhibiting EPHX1-Mediated Arachidonic Acid Metabolism
Abstract
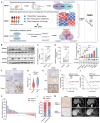
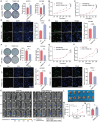
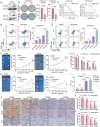
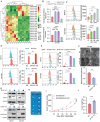
Pancreatic cancer (PC) progresses rapidly, and gemcitabine-based chemotherapy has brought only limited efficacy. Identifying key drivers and therapeutic targets holds significant clinical value. In this study, through comprehensive analysis of multiple PC databases, this work identifies TRIM21 as a promising driver mediator. This work further performs loss- and gain-of-function assays for TRIM21, revealing that TRIM21 knockout inhibits tumor proliferation and gemcitabine resistance both in vitro and vivo. Lipidomics reveal that silencing TRIM21 reduce the arachidonic acid production, and inhibit ferroptosis. Mechanically, through proteomics, ubiquitomics, and liquid chromatography-tandem mass spectrometry analysis, the key metabolic enzyme of arachidonic acid -EPHX1 is identified as a downstream substrate of TRIM21. TRIM21 interacts with EPHX1 through its SPRY domain and promotes ubiquitin-mediated degradation of EPHX1 via K33- and K48-linked ubiquitination at the K105 site. Given the targeting potential, this work screens Bezafibrate to block the interaction between TRIM21 and EPHX1 and validates its sensitizing effect. In summary, TRIM21 promotes tumour growth and gemcitabine resistance in PC by inhibiting EPHX1-mediated arachidonic acid metabolism. This provides a novel and promising target for clinical treatment of PC.
Keywords: EPHX1; TRIM21; arachidonic acid metabolism; pancreatic cancer; ubiquitination.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Siegel R. L., Miller K. D., Wagle N. S., Jemal A., Ca‐Cancer J. Clin. 2023, 73, 17. - PubMed
-
- Rahib L., Smith B. D., Aizenberg R., Rosenzweig A. B., Fleshman J. M., Matrisian L. M., Cancer Res. 2014, 74, 2913. - PubMed
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Cancer J. Clin. 2018, 68, 394. - PubMed
-
- Yu X., Holist. Integr. Oncol. 2024, 3, 35.
-
- Von Hoff D. D., Ervin T., Arena F. P., Chiorean E. G., Infante J., Moore M., Seay T., Tjulandin S. A., Ma W. W., Saleh M. N., Harris M., Reni M., Dowden S., Laheru D., Bahary N., Ramanathan R. K., Tabernero J., Hidalgo M., Goldstein D., Van Cutsem E., Wei X., Iglesias J., Renschler M. F., N. Engl. J. Med. 2013, 369, 1691. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
