Profiling endogenous airway proteases and antiproteases and modeling proteolytic activation of Influenza HA using in vitro and ex vivo human airway surface liquid samples
- PMID: 39739661
- PMCID: PMC11687774
- DOI: 10.1371/journal.pone.0306197
Profiling endogenous airway proteases and antiproteases and modeling proteolytic activation of Influenza HA using in vitro and ex vivo human airway surface liquid samples
Abstract
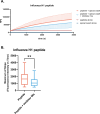
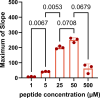
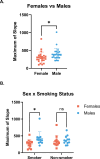
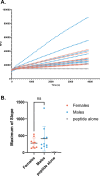
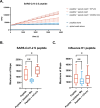
Imbalance of airway proteases and antiproteases has been implicated in diseases such as COPD and environmental exposures including cigarette smoke and ozone. To initiate infection, endogenous proteases are commandeered by respiratory viruses upon encountering the airway epithelium. The airway proteolytic environment likely contains redundant antiproteases and proteases with diverse catalytic mechanisms, however a proteomic profile of these enzymes and inhibitors in airway samples has not been reported. The objective of this study was to first profile extracellular proteases and antiproteases using human airway epithelial cell cultures and ex vivo nasal epithelial lining fluid (NELF) samples. Secondly, we present an optimized method for probing the proteolytic environment of airway surface liquid samples (in vitro and ex vivo) using fluorogenic peptides modeling the cleavage sites of respiratory viruses. We detected 48 proteases in the apical wash of cultured human nasal epithelial cells (HNECs) (n = 6) and 57 in NELF (n = 13) samples from healthy human subjects using mass-spectrometry based proteomics. Additionally, we detected 29 and 48 antiproteases in the HNEC apical washes and NELF, respectively. We observed large interindividual variability in rate of cleavage of an Influenza H1 peptide in the ex vivo clinical samples. Since protease and antiprotease levels have been found to be altered in the airways of smokers, we compared proteolytic cleavage in ex vivo nasal lavage samples from male/female smokers and non-smokers. There was a statistically significant increase in proteolysis of Influenza H1 in NLF from male smokers compared to female smokers. Furthermore, we measured cleavage of the S1/S2 site of SARS-CoV, SARS-CoV-2, and SARS-CoV-2 Delta peptides in various airway samples, suggesting the method could be used for other viruses of public health relevance. This assay presents a direct and efficient method of evaluating the proteolytic environment of human airway samples in assessment of therapeutic treatment, exposure, or underlying disease.
Copyright: © 2024 Brocke et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures.J Virol. 2020 Sep 15;94(19):e00985-20. doi: 10.1128/JVI.00985-20. Print 2020 Sep 15. J Virol. 2020. PMID: 32699094 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
References
-
- Antalis T.M., Buzza M.S. Extracellular: Plasma Membrane Proteases—Serine Proteases. Encyclopedia of Cell Biology. 2016:650–60.
-
- Sepper R, Konttinen YT, Ingman T, Sorsa T. Presence, activities, and molecular forms of cathepsin G, elastase,α1-antitrypsin, andα1-antichymotrypsin in bronchiectasis. Journal of Clinical Immunology. 1995;15(1):27–34. - PubMed
-
- S Yasuoka TO, Kawano S, Tsuchihashi S, Ogawara M, Masuda K, Yamaoka K, et al.. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. American Journal of Respiratory Cell and Molecular Biology. 1997;16(3):300–8. doi: 10.1165/ajrcmb.16.3.9070615 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

