Mechanism of connexin channel inhibition by mefloquine and 2-aminoethoxydiphenyl borate
- PMID: 39739741
- PMCID: PMC11687724
- DOI: 10.1371/journal.pone.0315510
Mechanism of connexin channel inhibition by mefloquine and 2-aminoethoxydiphenyl borate
Abstract
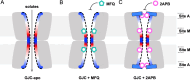
Gap junction intercellular communication (GJIC) between two adjacent cells involves direct exchange of cytosolic ions and small molecules via connexin gap junction channels (GJCs). Connexin GJCs have emerged as drug targets, with small molecule connexin inhibitors considered a viable therapeutic strategy in several diseases. The molecular mechanisms of GJC inhibition by known small molecule connexin inhibitors remain unknown, preventing the development of more potent and connexin-specific therapeutics. Here we show that two GJC inhibitors, mefloquine (MFQ) and 2-aminoethoxydiphenyl borate (2APB) bind to Cx32 and block dye permeation across Cx32 hemichannels (HCs) and GJCs. Cryo-EM analysis shows that 2APB binds to "site A", close to the N-terminal gating helix of Cx32 GJC, restricting the entrance to the channel pore. In contrast, MFQ binds to a distinct "site M", deeply buried within the pore. MFQ binding to this site modifies the electrostatic properties of Cx32 pore. Mutagenesis of V37, a key residue located in the site M, renders Cx32 HCs and GJCs insensitive to MFQ-mediated inhibition. Moreover, our cryo-EM analysis, mutagenesis and activity assays show that MFQ targets the M site in Cx43 GJC similarly to Cx32. Taken together, our results point to a conserved inhibitor binding site in connexin channels, opening a new route for development of specific drugs targeting connexins.
Copyright: © 2024 Lavriha et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Structure of the connexin-43 gap junction channel in a putative closed state.Elife. 2023 Aug 3;12:RP87616. doi: 10.7554/eLife.87616. Elife. 2023. PMID: 37535063 Free PMC article.
-
Mefloquine-induced conformational shift in Cx36 N-terminal helix leading to channel closure mediated by lipid bilayer.Nat Commun. 2024 Oct 25;15(1):9223. doi: 10.1038/s41467-024-53587-6. Nat Commun. 2024. PMID: 39455592 Free PMC article.
-
Emerging issues of connexin channels: biophysics fills the gap.Q Rev Biophys. 2001 Aug;34(3):325-472. doi: 10.1017/s0033583501003705. Q Rev Biophys. 2001. PMID: 11838236 Review.
-
Structures of wild-type and selected CMT1X mutant connexin 32 gap junction channels and hemichannels.Sci Adv. 2023 Sep;9(35):eadh4890. doi: 10.1126/sciadv.adh4890. Epub 2023 Aug 30. Sci Adv. 2023. PMID: 37647412 Free PMC article.
-
Therapeutic Targeting of Connexin Channels: New Views and Challenges.Trends Mol Med. 2018 Dec;24(12):1036-1053. doi: 10.1016/j.molmed.2018.10.005. Epub 2018 Nov 10. Trends Mol Med. 2018. PMID: 30424929 Review.
References
-
- Totland MZ, Rasmussen NL, Knudsen LM, Leithe E. Regulation of gap junction intercellular communication by connexin ubiquitination: physiological and pathophysiological implications. Cell Mol Life Sci. 2020;77(4):573–91. Epub 20190909. doi: 10.1007/s00018-019-03285-0 ; PubMed Central PMCID: PMC7040059. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

