Topological confinement by a membrane anchor suppresses phase separation into protein aggregates: Implications for prion diseases
- PMID: 39739794
- PMCID: PMC11725851
- DOI: 10.1073/pnas.2415250121
Topological confinement by a membrane anchor suppresses phase separation into protein aggregates: Implications for prion diseases
Abstract
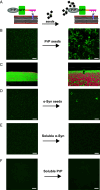
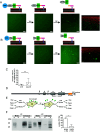
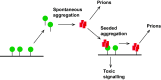
Protein misfolding and aggregation are a hallmark of various neurodegenerative disorders. However, the underlying mechanisms driving protein misfolding in the cellular context are incompletely understood. Here, we show that the two-dimensional confinement imposed by a membrane anchor stabilizes the native protein conformation and suppresses liquid-liquid phase separation (LLPS) and protein aggregation. Inherited prion diseases in humans and neurodegeneration in transgenic mice are linked to the expression of anchorless prion protein (PrP), suggesting that the C-terminal glycosylphosphatidylinositol (GPI) anchor of native PrP impedes spontaneous formation of neurotoxic and infectious PrP species. Combining unique in vitro and in vivo approaches, we demonstrate that anchoring to membranes prevents LLPS and spontaneous aggregation of PrP. Upon release from the membrane, PrP undergoes a conformational transition to detergent-insoluble aggregates. Our study demonstrates an essential role of the GPI anchor in preventing spontaneous misfolding of PrPC and provides a mechanistic basis for inherited prion diseases associated with anchorless PrP.
Keywords: liquid-liquid phase separation; membrane; neurodegenerative diseases; prion; protein aggregation.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Similar articles
-
A comprehensive phylogeny of mammalian PRNP gene reveals no influence of prion misfolding propensity on the evolution of this gene.PLoS Pathog. 2025 Jun 25;21(6):e1013257. doi: 10.1371/journal.ppat.1013257. eCollection 2025 Jun. PLoS Pathog. 2025. PMID: 40561181 Free PMC article.
-
Effect of primary structural variation on cervid prion protein in flexibility, stability, and spontaneous misfolding propensity.Neurobiol Dis. 2025 Sep;213:107005. doi: 10.1016/j.nbd.2025.107005. Epub 2025 Jun 13. Neurobiol Dis. 2025. PMID: 40517962
-
Prion protein facilitates retinal iron uptake and is cleaved at the β-site: Implications for retinal iron homeostasis in prion disorders.Sci Rep. 2017 Aug 29;7(1):9600. doi: 10.1038/s41598-017-08821-1. Sci Rep. 2017. PMID: 28851903 Free PMC article.
-
The infectious potential of protein misfolding: insights from cross-over studies of prion diseases and common neurodegenerative disorders.Microb Pathog. 2025 Sep;206:107789. doi: 10.1016/j.micpath.2025.107789. Epub 2025 Jun 4. Microb Pathog. 2025. PMID: 40480448 Review.
-
A review on current theories and potential therapies for prion diseases.Mol Biol Rep. 2025 Jul 4;52(1):674. doi: 10.1007/s11033-025-10754-2. Mol Biol Rep. 2025. PMID: 40613913 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

