RAP-2 and CNH-MAP4 Kinase MIG-15 confer resistance in bystander epithelium to cell-fate transformation by excess Ras or Notch activity
- PMID: 39739816
- PMCID: PMC11725784
- DOI: 10.1073/pnas.2414321121
RAP-2 and CNH-MAP4 Kinase MIG-15 confer resistance in bystander epithelium to cell-fate transformation by excess Ras or Notch activity
Abstract
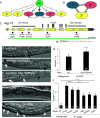
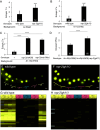
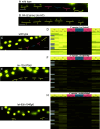
Induction of cell fates by growth factors impacts many facets of developmental biology and disease. LIN-3/EGF induces the equipotent vulval precursor cells (VPCs) in Caenorhabditis elegans to assume the 3˚-3˚-2˚-1˚-2˚-3˚ pattern of cell fates. 1˚ and 2˚ cells become specialized epithelia and undergo stereotyped series of cell divisions to form the vulva. Conversely, 3˚ cells are relatively quiescent and nonspecialized; they divide once and fuse with the surrounding epithelium. 3˚ cells have thus been characterized as passive, uninduced, or ground state. Based on our previous studies, we hypothesized that a 3˚-promoting program would confer resistance to cell fate-transformation by inappropriately activated 1˚ and 2˚ fate-promoting LET-60/Ras and LIN-12/Notch, respectively. Deficient MIG-15/CNH-MAP4 Kinase meets the expectations of genetic interactions for a 3˚-promoting protein. Moreover, endogenous MIG-15 is required for expression of a fluorescent biomarker of 3˚ cell fate, is expressed in VPCs, and functions cell autonomously in VPCs. The Ras family small GTPase RAP-2, orthologs of which activate orthologs of MIG-15 in other systems, emulates these functions of MIG-15. However, gain of RAP-2 function has no effect on patterning, suggesting its activity is constitutive in VPCs. The 3˚ biomarker is expressed independently of the AC, raising questions about the cellular origin of 3˚-promoting activity. Activated LET-60/Ras and LIN-12/Notch repress expression of the 3˚ biomarker, suggesting that the 3˚-promoting program is both antagonized by as well as antagonizes 1˚- and 2˚- promoting programs. This study provides insight into developmental properties of cells historically considered to be nonresponding to growth factor signals.
Keywords: ACDS-10; LET-60; LIN-12; Rap2; citron NIK1 homology.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

