An immunoinformatics and extensive molecular dynamics study to develop a polyvalent multi-epitope vaccine against cryptococcosis
- PMID: 39739919
- PMCID: PMC11687922
- DOI: 10.1371/journal.pone.0315105
An immunoinformatics and extensive molecular dynamics study to develop a polyvalent multi-epitope vaccine against cryptococcosis
Retraction in
-
Retraction: An immunoinformatics and extensive molecular dynamics study to develop a polyvalent multi-epitope vaccine against cryptococcosis.PLoS One. 2025 Apr 4;20(4):e0322316. doi: 10.1371/journal.pone.0322316. eCollection 2025. PLoS One. 2025. PMID: 40184336 Free PMC article. No abstract available.
Abstract
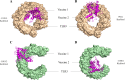
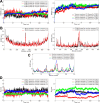
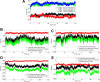
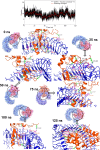
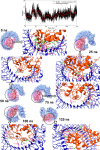
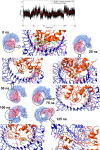
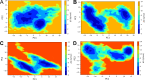
Cryptococcosis is a lethal mycosis instigated by the pathogenic species Cryptococcus neoformans and Cryptococcus gattii, primarily affects the lungs, manifesting as pneumonia, and the brain, where it presents as meningitis. Mortality rate could reach 100% if infections remain untreated in cryptococcal meningitis. Treatment options for cryptococcosis are limited and and there are no licensed vaccines clinically available to treat or prevent cryptococcosis. Our study utilizes an integrated bioinformatics approaches to develop a polyvalent multiepitope subunit vaccine focusing on the key virulent proteins Heat shock transcription factor and Chaperone DnaK of both C. neoformans and C. gatti. Then in silico analysis was done to predict highly antigenic epitopes by assessing antigenicity, transmembrane topology screening, allergenecity, toxicity, and molecular docking approaches. Following this analysis, we designed two vaccine constructs integrating a compatible adjuvant and suitable linkers. These constructs exhibited notable characteristics including high antigenicity, non-toxicity, solubility, stability, and compatibility with Toll-like receptors (TLRs). The interaction between both vaccine constructs and TLR2, TLR3, and TLR9 was assessed through molecular docking analysis. Molecular dynamics simulations and MM-PBSA calculations suggest the substantial stabilizing property and binding affinity of Vaccine Construct V1 against TLR9. Both the vaccines revealed to have a higher number of interchain hydrogen bond with TLR9. These findings serve as a crucial stepping stone towards a comprehensive solution for combating cryptococcus infections induced by both C. neoformans and C. gattii. Further validation through in vivo studies is crucial to confirm the effectiveness and potential of the vaccine to curb the spread of cryptococcosis. Subsequent validation through in vivo studies is paramount to confirm the effectiveness and potential of the vaccine in reducing the spread of cryptococcosis.
Copyright: © 2024 Sami et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Firacative C, Meyer W, Castañeda E. Cryptococcus neoformans and Cryptococcus gattii species complexes in Latin America: a map of molecular types, genotypic diversity, and antifungal susceptibility as reported by the Latin American Cryptococcal Study Group. Journal of Fungi. 2021;7(4):282. doi: 10.3390/jof7040282 - DOI - PMC - PubMed
-
- Hansakon A, Mutthakalin P, Ngamskulrungroj P, Chayakulkeeree M, Angkasekwinai P. Cryptococcus neoformans and Cryptococcus gattii clinical isolates from Thailand display diverse phenotypic interactions with macrophages. Virulence. 2019;10(1):26–36. doi: 10.1080/21505594.2018.1556150 - DOI - PMC - PubMed
-
- Kenosi K, Mosimanegape J, Daniel L, Ishmael K. Recent Advances in the Ecoepidemiology, Virulence and Diagnosis of Cryptococcus neoformans and Cryptococcus gattii Species Complexes. The Open Microbiology Journal. 2023;17(1).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

