miRNAome-metabolome wide association study reveals effects of miRNA regulation in eosinophilia and airflow obstruction in childhood asthma
- PMID: 39740296
- PMCID: PMC11750448
- DOI: 10.1016/j.ebiom.2024.105534
miRNAome-metabolome wide association study reveals effects of miRNA regulation in eosinophilia and airflow obstruction in childhood asthma
Abstract
Background: There are important inter-relationships between miRNAs and metabolites: alterations in miRNA expression can be induced by various metabolic stimuli, and miRNAs play a regulatory role in numerous cellular processes, impacting metabolism. While both specific miRNAs and metabolites have been identified for their role in childhood asthma, there has been no global assessment of the combined effect of miRNAs and the metabolome in childhood asthma.
Methods: We performed miRNAome-metabolome-wide association studies ('miR-metabo-WAS') in two childhood cohorts of asthma to evaluate the contemporaneous and persistent miRNA-metabolite associations: 1) Genetic Epidemiology of Asthma in Costa Rica Study (GACRS) (N = 1121); 2) the Childhood Asthma Management Program (CAMP) (NBaseline = 312 and NEnd of trial = 454). We conducted a meta-analysis of the two cohorts to identify common contemporaneous associations between CAMP and GACRS (false-discovery rate (FDR) = 0.05). We assessed persistent miRNA-metabolome associations using baseline miRNAs and metabolomic profiling in CAMP at the end of the trial. The relation between miRNAs, metabolites and clinical phenotypes, including airway hyper-responsiveness (AHR), peripheral blood eosinophilia, and airflow obstruction, were then assessed via. Mediation analysis with 1000 bootstraps at an FDR significance level of 0.05.
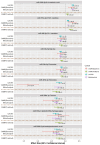
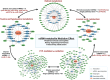
Findings: The meta-analysis yielded a total of 369 significant contemporaneous associations, involving 133 miRNAs and 60 metabolites. We identified 13 central hub metabolites (taurine, 12,13-diHOME, sebacate, 9-cis-retinoic acid, azelate, asparagine, C5:1 carnitine, cortisol, 3-methyladipate, inosine, NMMA, glycine, and Pyroglutamic acid) and four hub miRNAs (hsa-miR-186-5p, hsa-miR-143-3p, hsa-miR-192-5p, and hsa-miR-223-3p). Nine of these associations, between eight miRNAs and eight metabolites, were persistent in CAMP from baseline to the end of trial. Finally, five central hub metabolites (9-cis-retinoic acid, taurine, sebacate, azelate, and 12,13-diHOME) were identified as primary mediators in over 100 significant indirect miRNA-metabolite associations, with a collective influence on peripheral blood eosinophilia, AHR, and airflow obstruction.
Interpretation: The robust association between miRNAs and metabolites, along with the substantial indirect impact of miRNAs via 5 hub metabolites on multiple clinical asthma metrics, suggests important integrated effects of miRNAs and metabolites on asthma. These findings imply that the indirect regulation of metabolism and cellular functions by miRNA influences Th2 inflammation, AHR, and airflow obstruction in childhood asthma.
Funding: Molecular data for CAMP and GACRS via the Trans-Omics in Precision Medicine (TOPMed) program was supported by the National Heart, Lung, and Blood Institute (NHLBI).
Keywords: Airflow obstruction; Airway hyper-responsiveness; Eosinophilia; Metabolite; microRNA.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests JLS is a scientific advisor to Precion, Inc., receives grants and consulting fees from TruDiagnostic and Ahara Corp, and holds patents with TruDiagnostic. STW is a board member of Histolix and receives royalties from UpToDate. All other authors declare no potential, perceived, or real conflict of interest regarding the content of this manuscript.
Figures


References
-
- Papi A., Brightling C., Pedersen S.E., Reddel H.K. Asthma. Lancet (London, England) 2018;391(10122):783–800. - PubMed
-
- Porsbjerg C., Melén E., Lehtimäki L., Shaw D. Asthma. Lancet (London, England) 2023;401(10379):858–873. - PubMed
-
- Services 2019 national health interview survey data. U.S. Department of health & human. Centers for Disease Control and Prevention; 2020. https://www.cdc.gov/asthma/nhis/2019/data.htm Available from:
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL169300/HL/NHLBI NIH HHS/United States
- K01 HL153941/HL/NHLBI NIH HHS/United States
- R01 HL155742/HL/NHLBI NIH HHS/United States
- R01 HL141826/HL/NHLBI NIH HHS/United States
- R01 HL139634/HL/NHLBI NIH HHS/United States
- HHSN268201800001C/HL/NHLBI NIH HHS/United States
- U01 HG008685/HG/NHGRI NIH HHS/United States
- R01 HL127332/HL/NHLBI NIH HHS/United States
- R01 HL123915/HL/NHLBI NIH HHS/United States
- R01 HL120393/HL/NHLBI NIH HHS/United States
- U01 HL120393/HL/NHLBI NIH HHS/United States
- U19 AI168643/AI/NIAID NIH HHS/United States
- P01 HL132825/HL/NHLBI NIH HHS/United States
- K01 HL146980/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

