Rapid detection of Pan-Avian Influenza Virus and H5, H7, H9 subtypes of Avian Influenza Virus using CRISPR/Cas13a and lateral flow assay
- PMID: 39740498
- PMCID: PMC11750554
- DOI: 10.1016/j.psj.2024.104745
Rapid detection of Pan-Avian Influenza Virus and H5, H7, H9 subtypes of Avian Influenza Virus using CRISPR/Cas13a and lateral flow assay
Abstract
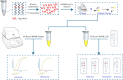
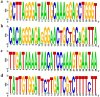
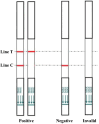
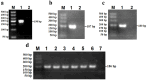
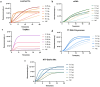
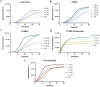
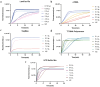
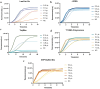
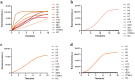
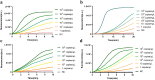
Avian Influenza Virus (AIV) has been prevalent worldwide in recent years, resulting in substantial economic losses in the poultry industry. More importantly, AIV is capable of cross-species transmission among mammals, posing a dormant yet considerable threat to human health and safety. In this study, two rapid detection methods for AIV based on the CRISPR-Cas13a were developed. These methods can identify AIV through the M gene and differentiate the H5, H7, and H9 subtypes via the HA gene. The first method utilizes RT-RAA isothermal amplification of the target sequence in combination with the "collateral effect" of the Cas13a protein. The results are measured using a real-time quantitative PCR instrument, with a Limit of Detection (LOD) as low as 1 copy/μL. The second method combines RT-RAA with Cas13a and a lateral flow assay, allowing results to be visually observed with the naked eye, with a LOD of 10 copies/μL. Both methods demonstrated specificity and sensitivity comparable to or exceeding that of qRT-PCR, suggesting strong potential for clinical application.
Keywords: Avian influenza virus; CRISPR/Cas13a; Lateral flow assay; RT-RAA; Rapid detection.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare no conflicts of interest.
Figures

References
-
- Barnes K.G., Lachenauer A.E., Nitido A., Siddiqui S., Gross R., Beitzel B., Siddle K.J., Freije C.A., Dighero-Kemp B., Mehta S.B., Carter A., Uwanibe J., Ajogbasile F., Olumade T., Odia I., Sandi J.D., Momoh M., Metsky H.C., Boehm C.K., Lin A.E., Kemball M., Park D.J., Branco L., Boisen M., Sullivan B., Amare M.F., Tiamiyu A.B., Parker Z.F., Iroezindu M., Grant D.S., Modjarrad K., Myhrvold C., Garry R.F., Palacios G., Hensley L.E., Schaffner S.F., Happi C.T., Colubri A., Sabeti P.C. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat. Commun. 2020;11:4131. - PMC - PubMed
-
- Bi Y., Li J., Li S., Fu G., Jin T., Zhang C., Yang Y., Ma Z., Tian W., Li J., Xiao S., Li L., Yin R., Zhang Y., Wang L., Qin Y., Yao Z., Meng F., Hu D., Li D., Wong G., Liu F., Lv N., Wang L., Fu L., Yang Y., Peng Y., Ma J., Sharshov K., Shestopalov A., Gulyaeva M., Gao G.F., Chen J., Shi Y., Liu W.J., Chu D., Huang Y., Liu Y., Liu L., Liu W., Chen Q., Shi W. Dominant subtype switch in avian influenza viruses during 2016–2019 in China. Nat. Commun. 2020;11:5909. - PMC - PubMed
-
- Chen W., Yu J., Xu H., Lu X., Dai T., Tian Y., Shen D., Dou D. Combining simplified DNA extraction technology and recombinase polymerase amplification assay for rapid and equipment-free detection of citrus pathogen Phytophthora parasitica. J. Integr. Agric. 2021;20:2696–2705.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

