Evaluation of a Decentralized Donor-Derived Cell-Free DNA Assay for Kidney Allograft Rejection Monitoring
- PMID: 39741495
- PMCID: PMC11685011
- DOI: 10.3389/ti.2024.13919
Evaluation of a Decentralized Donor-Derived Cell-Free DNA Assay for Kidney Allograft Rejection Monitoring
Abstract
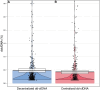
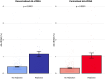
Donor-derived cell-free DNA (dd-cfDNA) is an emerging non-invasive biomarker for allograft injury detection. This study aimed to evaluate a new, decentralized dd-cfDNA testing kit against a centralized dd-cfDNA testing service broadly utilized in the United States. Kidney transplant recipients with decentralized and centralized dd-cfDNA measurements and concomitant kidney allograft biopsies were included in the study. 580 kidney allograft recipients from 3 referral centers were included for 603 total evaluations. Correlation between assays was evaluated using r-squared (r 2) and Spearman's rank correlation test, and associations with rejection using logistic regression analyses and discrimination using area under the curve. Mean dd-cfDNA levels from decentralized and centralized tests were 0.51% ± 0.81% and 0.43% ± 0.78%, respectively. The assays were highly correlated, with r 2 = 0.95 and Spearman's rank correlation 0.88 (p < 0.0001). Both tests showed significant association with allograft rejection (p < 0.0001) and good and similar discriminations to predict rejection (AUC: 0.758 for the decentralized and AUC: 0.760 for the centralized dd-cfDNA; p = 0.8466). Consistency between the assays was also confirmed across clinical scenarios including post-transplant timepoint, allograft stability, and allograft rejection subcategories. This decentralized dd-cfDNA assessment demonstrates high accuracy and value to non-invasively monitor kidney recipients.
Keywords: AlloSeq; allograft rejection; dd-cfDNA; liquid biopsy; non-invasive diagnosis.
Copyright © 2024 Loupy, Certain, Tangprasertchai, Racapé, Ursule-Dufait, Benbadi, Raynaud, Vaskova, Marchis, Casas, Hague, Bestard, Kervella, Lefaucheur, Viard and Aubert.
Conflict of interest statement
CareDx participated in dd-cfDNA testing by providing reagents for decentralized testing, blinded to clinical information, and reviewed the manuscript and data analysis. Authors NT, EV, CM, SC, TH, and TV were employed by the company CareDx. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

