Evaluation of intestinal biopsy tissue preservation methods to facilitate large-scale mucosal microbiota research
- PMID: 39742562
- PMCID: PMC11751561
- DOI: 10.1016/j.ebiom.2024.105550
Evaluation of intestinal biopsy tissue preservation methods to facilitate large-scale mucosal microbiota research
Abstract
Background: Large-scale multicentre studies are needed to understand complex relationships between the gut microbiota, health and disease. Interrogating the mucosal microbiota may identify important biology not captured by stool analysis. Gold standard tissue cryopreservation ('flash freezing') limits large-scale study feasibility. We aimed to compare gut microbiota in gold standard and pragmatic mucosal biopsy storage conditions.
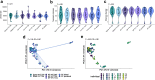
Methods: We collected endoscopic recto-sigmoid biopsies from 20 adults with inflammatory bowel disease. Biopsies were preserved using three methods: (i) flash freezing (most proximal and distal biopsy sites); (ii) nucleic acid preservative reagents (QIAGEN Allprotect®, Invitrogen RNAlater™, and Zymo DNA/RNA Shield™); and (iii) formalin fixation with paraffin embedding (FFPE), which is used to preserve tissue for clinical histopathology within healthcare settings. Microbiota were sequenced on the MiSeq platform (V4 region, 16S rRNA gene).
Findings: Tissue microbiota were consistent between most proximal and distal tissue suggesting any within-patient variation observed reflected storage condition, not biopsy location. There was no significant difference in alpha-diversity or microbial community profiles of reagent-preserved versus gold standard tissue. FFPE community structure was significantly dissimilar to other tissue samples, driven by differential relative abundance of obligate gut anaerobes; Faecalibacterium, Anaerostipes and Lachnospiraceae. Despite these differences, tissue microbiota grouped by participant regardless of preservation and storage conditions.
Interpretation: Preservative reagents offer a convenient alternative to flash freezing tissue in prospective large-scale mucosal microbiota studies. Whilst less comparable, FFPE provides potential for retrospective microbiota studies using historical samples.
Funding: Medical Research Council (MR/T032162/1) and The Leona M. and Harry B. Helmsley Charitable Trust (G-2002-04255).
Keywords: Formalin fixed paraffin embedded; Gut microbiome; Inflammatory bowel disease; Precision medicine; Tissue microbiome; Tissue preservative reagents.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests None to declare.
Figures


References
-
- Riedel C.U., Schwiertz A., Egert M. In: The human microbiota and microbiome. Marchesi J.R., editor. CAB International; Oxfordshire, UK: 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

