Operating principles of interconnected feedback loops driving cell fate transitions
- PMID: 39743534
- PMCID: PMC11693754
- DOI: 10.1038/s41540-024-00483-w
Operating principles of interconnected feedback loops driving cell fate transitions
Abstract
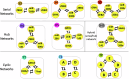
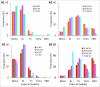
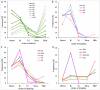
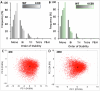
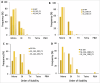
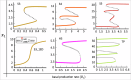
Interconnected feedback loops are prevalent across biological mechanisms, including cell fate transitions enabled by epigenetic mechanisms in carcinomas. However, the operating principles of these networks remain largely unexplored. Here, we identify numerous interconnected feedback loops implicated in cell lineage decisions, which we discover to be the hallmarks of lower- and higher-dimensional state space. We demonstrate that networks having higher centrality nodes have restricted state space while those with lower centrality nodes have higher dimensional state space. The topologically distinct networks with identical node or loop counts have different steady-state distributions, highlighting the crucial influence of network structure on emergent dynamics. Further, regardless of topology, networks with autoregulated nodes exhibit multiple steady states, thereby "liberating" network dynamics from absolute topological control. These findings unravel the design principles of multistable networks implicated in fate decisions and can have crucial implications in engineering or comprehending multi-fate decision circuits.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Moris, N., Pina, C. & Martinez Arias, A. Transition states and cell fate decisions in epigenetic landscapes. 10.1038/nrg.2016.98 (2016). - PubMed
-
- Zhou, J. X. & Huang, S. In press. Understanding gene circuits at cell-fate branch points for rational cell reprogramming. 10.1016/j.tig.2010.11.002 - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

