Monoallelic expression can govern penetrance of inborn errors of immunity
- PMID: 39743591
- PMCID: PMC11804961
- DOI: 10.1038/s41586-024-08346-4
Monoallelic expression can govern penetrance of inborn errors of immunity
Abstract
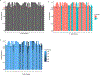
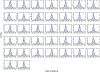
Inborn errors of immunity (IEIs) are genetic disorders that underlie susceptibility to infection, autoimmunity, autoinflammation, allergy and/or malignancy1. Incomplete penetrance is common among IEIs despite their monogenic basis2. Here we investigate the contribution of autosomal random monoallelic expression (aRMAE), a somatic commitment to the expression of one allele3,4, to phenotypic variability observed in families with IEIs. Using a clonal primary T cell system to assess aRMAE status of genes in healthy individuals, we find that 4.30% of IEI genes and 5.20% of all genes undergo aRMAE. Perturbing H3K27me3 and DNA methylation alters allele expression commitment, in support of two proposed mechanisms5,6 for the regulation of aRMAE. We tested peripheral blood mononuclear cells from individuals with IEIs with shared genetic lesions but discordant clinical phenotypes for aRMAE. Among two relatives who were heterozygous for a mutation in PLCG2 (delEx19), an antibody deficiency phenotype corresponds to selective mutant allele expression in B cells. By contrast, among relatives who were heterozygous for a mutation in JAK1 (c.2099G>A; p.S700N), the unaffected carrier T cells predominantly expressed the wild-type JAK1 allele, whereas the affected carrier T cells exhibited biallelic expression. Allelic expression bias was also documented in phenotypically discordant family members with mutations in STAT1 and CARD11. This study highlights the importance of considering both the genotype and the 'transcriptotype' in analyses of the penetrance and expressivity of monogenic disorders.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: D.B. is the founder of Lab11 Therapeutics. The other authors declare no competing interests.
Figures
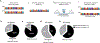


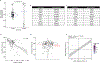









References
MeSH terms
Substances
Grants and funding
- R01 AI127372/AI/NIAID NIH HHS/United States
- R01 AI120989/AI/NIAID NIH HHS/United States
- R01 AI148963/AI/NIAID NIH HHS/United States
- R01 AI095983/AI/NIAID NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- T32 AI007512/AI/NIAID NIH HHS/United States
- R35 NS105078/NS/NINDS NIH HHS/United States
- U19 AI162568/AI/NIAID NIH HHS/United States
- F31 AI174808/AI/NIAID NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R01 AI127564/AI/NIAID NIH HHS/United States
- T32 HD075735/HD/NICHD NIH HHS/United States
- R24 AI167802/AI/NIAID NIH HHS/United States
- T32 AI078892/AI/NIAID NIH HHS/United States
- R01 AI151029/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

