Brain-wide cell-type-specific transcriptomic signatures of healthy ageing in mice
- PMID: 39743592
- PMCID: PMC11798837
- DOI: 10.1038/s41586-024-08350-8
Brain-wide cell-type-specific transcriptomic signatures of healthy ageing in mice
Abstract
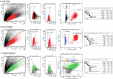
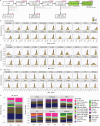
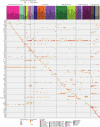
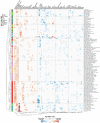
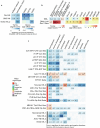
Biological ageing can be defined as a gradual loss of homeostasis across various aspects of molecular and cellular function1,2. Mammalian brains consist of thousands of cell types3, which may be differentially susceptible or resilient to ageing. Here we present a comprehensive single-cell RNA sequencing dataset containing roughly 1.2 million high-quality single-cell transcriptomes of brain cells from young adult and aged mice of both sexes, from regions spanning the forebrain, midbrain and hindbrain. High-resolution clustering of all cells results in 847 cell clusters and reveals at least 14 age-biased clusters that are mostly glial types. At the broader cell subclass and supertype levels, we find age-associated gene expression signatures and provide a list of 2,449 unique differentially expressed genes (age-DE genes) for many neuronal and non-neuronal cell types. Whereas most age-DE genes are unique to specific cell types, we observe common signatures with ageing across cell types, including a decrease in expression of genes related to neuronal structure and function in many neuron types, major astrocyte types and mature oligodendrocytes, and an increase in expression of genes related to immune function, antigen presentation, inflammation, and cell motility in immune cell types and some vascular cell types. Finally, we observe that some of the cell types that demonstrate the greatest sensitivity to ageing are concentrated around the third ventricle in the hypothalamus, including tanycytes, ependymal cells, and certain neuron types in the arcuate nucleus, dorsomedial nucleus and paraventricular nucleus that express genes canonically related to energy homeostasis. Many of these types demonstrate both a decrease in neuronal function and an increase in immune response. These findings suggest that the third ventricle in the hypothalamus may be a hub for ageing in the mouse brain. Overall, this study systematically delineates a dynamic landscape of cell-type-specific transcriptomic changes in the brain associated with normal ageing that will serve as a foundation for the investigation of functional changes in ageing and the interaction of ageing and disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: H.Z. is on the scientific advisory board of MapLight Therapeutics, Inc. The other authors declare no competing interests.
Figures















References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

