Dysregulation of mTOR signalling is a converging mechanism in lissencephaly
- PMID: 39743596
- PMCID: PMC11798849
- DOI: 10.1038/s41586-024-08341-9
Dysregulation of mTOR signalling is a converging mechanism in lissencephaly
Abstract
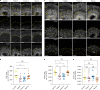
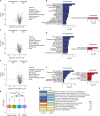
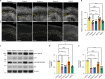
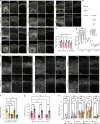
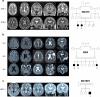
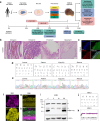
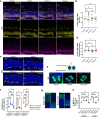
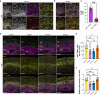
Cerebral cortex development in humans is a highly complex and orchestrated process that is under tight genetic regulation. Rare mutations that alter gene expression or function can disrupt the structure of the cerebral cortex, resulting in a range of neurological conditions1. Lissencephaly ('smooth brain') spectrum disorders comprise a group of rare, genetically heterogeneous congenital brain malformations commonly associated with epilepsy and intellectual disability2. However, the molecular mechanisms underlying disease pathogenesis remain unknown. Here we establish hypoactivity of the mTOR pathway as a clinically relevant molecular mechanism in lissencephaly spectrum disorders. We characterized two types of cerebral organoid derived from individuals with genetically distinct lissencephalies with a recessive mutation in p53-induced death domain protein 1 (PIDD1) or a heterozygous chromosome 17p13.3 microdeletion leading to Miller-Dieker lissencephaly syndrome (MDLS). PIDD1-mutant organoids and MDLS organoids recapitulated the thickened cortex typical of human lissencephaly and demonstrated dysregulation of protein translation, metabolism and the mTOR pathway. A brain-selective activator of mTOR complex 1 prevented and reversed cellular and molecular defects in the lissencephaly organoids. Our findings show that a converging molecular mechanism contributes to two genetically distinct lissencephaly spectrum disorders.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.S. is a co-founder and shareholder of Bexorg, Inc. All the other authors declare no competing interests.
Figures








References
MeSH terms
Substances
Grants and funding
- R01 HD110693/HD/NICHD NIH HHS/United States
- UM1 MH130991/MH/NIMH NIH HHS/United States
- UM1 HG006504/HG/NHGRI NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 MH122678/MH/NIMH NIH HHS/United States
- U01 MH122678/MH/NIMH NIH HHS/United States
- S10 OD018034/OD/NIH HHS/United States
- RC2 NS070477/NS/NINDS NIH HHS/United States
- S10 OD019967/OD/NIH HHS/United States
- U01 DA053628/DA/NIDA NIH HHS/United States
- U01 MH116488/MH/NIMH NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
- U01 MH124619/MH/NIMH NIH HHS/United States
- R01 MH103616/MH/NIMH NIH HHS/United States
- S10 OD030363/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

