HPV-Associated Gene Signatures in Bladder Cancer: A Comprehensive Prognostic Model and its Implications in Immunotherapy
- PMID: 39744172
- PMCID: PMC11659835
- DOI: 10.7150/ijms.98334
HPV-Associated Gene Signatures in Bladder Cancer: A Comprehensive Prognostic Model and its Implications in Immunotherapy
Abstract
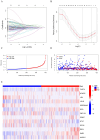
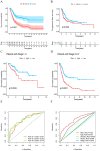
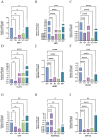
Background: Evidence increasingly indicates that HPV infection plays a pivotal role in the initiation and progression of bladder cancer (BC). Yet, determining the predictive value of HPV-associated genes in BC remains challenging. Methods: We identified differentially expressed HPV-associated genes of BC patients from the TCGA and GEO databases. We screened prognostic genes using COX and LASSO regression, subsequently establishing a risk prediction model. The model's precision and clinical relevance were gauged using Kaplan-Meier survival analyses and ROC curves. Functional enrichment, immune cell infiltration, and drug sensitivity analyses were performed across both high-risk and low-risk sets. PCR assays were utilized to measure the expression levels of genes. Results: We identified 13 HPV-associated genes for our risk model. Among these, FLRT2, HOXC5, LDLR, SCD, GRM7, DSC1, EMP1, and HMGA1 were identified as risk contributors, while LPA, SERPINA6, ZNF124, ETV7, and SCO2 were deemed protective. Cox regression analysis verified that our model provides an independent prediction of overall survival (OS) in bladder cancer (BC) patients. Gene Ontology (GO) analysis revealed predominant gene enrichment in wound healing, extracellular matrix composition, and collagen-rich extracellular matrices. KEGG pathway analysis highlighted primary enrichment areas, including focal adhesion, the PI3K-Akt signalling pathway, and ECM-receptor interaction. Risk scores were correlated with tumor microenvironment (TME) scores, immune cell infiltration, and sensitivities to both chemotherapy and immunotherapy. Conclusion: We have formulated a risk-assessment model pinpointing 13 central HPV-associated genes in BC. These genes present potential as prognostic indicators and therapeutic targets, emphasizing the intertwined relationship between HPV-induced BC progression and the immune landscape.
Keywords: Bladder cancer; Human papillomavirus (HPV); Prognostic model.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures




References
-
- Lobo N, Afferi L, Moschini M, Mostafid H, Porten S, Psutka SP. et al. Epidemiology, Screening, and Prevention of Bladder Cancer. Eur Urol Oncol. 2022 Dec;5(6):628–39. - PubMed
-
- Laaksonen MA, MacInnis RJ, Canfell K, Giles GG, Hull P, Shaw JE. et al. The future burden of kidney and bladder cancers preventable by behavior modification in Australia: A pooled cohort study. Int J Cancer. 2020 Feb;146(3):874–83. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

