A STING agonist prodrug reprograms tumor-associated macrophage to boost colorectal cancer immunotherapy
- PMID: 39744236
- PMCID: PMC11667223
- DOI: 10.7150/thno.101001
A STING agonist prodrug reprograms tumor-associated macrophage to boost colorectal cancer immunotherapy
Abstract
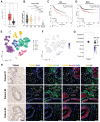
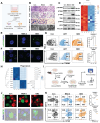
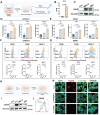
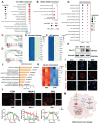
Rationale: Tumor-associated macrophages (TAMs) are abundant in colorectal cancer (CRC), correlating with immunosuppression and disease progression. Activation of the stimulator of interferon gene (STING) signaling pathway in TAMs offers a promising approach for CRC therapy. However, current STING agonists face challenges related to tumor specificity and administration routes. Method: The Cancer Genome Atlas (TCGA) database analysis and multicolor immunofluorescence experiments of human CRC samples analysed triggering receptor expressed on myeloid cells 2 (TREM2) expression in the tumor microenvironment of CRC patients. We designed and synthesized a STING agonist prodrug GB2 to reprogram TAMs by targeting TREM2 in tumors. Preliminary evaluation of the anti-tumor capacity of prodrug GB2 in the mouse CRC model intravenously. RNA-seq analysis of bone marrow-derived macrophages (BMDM) after GB2 treatment reveals novel pharmacological mechanisms for the prodrug GB2. Results: Over-expressed TREM2 in TAMs correlates with CRC progression. Via targeting TREM2 expressed in TAMs, GB2 induces comprehensive tumor regression by administrating intravenously in mouse colon cancer models, as well as in a STINGlow mouse melanoma model, with no systemic toxicity. Upon treatment with GB2, TAMs exhibit an M1 phenotype with pro-inflammatory function and demonstrate enhanced phagocytosis capacity. The molecular mechanisms involve (1) GB2 upregulating the Glycolysis-ROS-HIF-1α axis, thereby promoting glucose metabolism and inflammatory cytokine expression; (2) GB2 inducing endoplasmic reticulum-mitochondria contact (MERC), leading to mitochondrial fission, ultimately facilitating Ca2+-mediated phagocytosis. Besides, GB2-treated macrophages reverse immunosuppression, facilitating CD8+ T cell tumor infiltration and effector function. Combining GB2 with αPD-1 therapy reveals a synergistic effect on tumor inhibition, leading to prolonged mouse survival. Conclusion: By targeting TREM2 and activating the STING signaling pathway in TAMs, prodrug GB2 exhibits excellent anti-tumor efficacy and immune-activating capacity in the mouse colon cancer model.
Keywords: STING agonist; TREM2 inhibitor; colorectal cancer; prodrug; tumor associated macrophage.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Horesh N, Emile SH, Freund M, Garoufalia Z, Gefen R, Zhou PG. et al. Immunotherapy in locally advanced and metastatic rectal cancer: A propensity score matched analysis of the National Cancer Database. J Clin Oncol. 2023;41:e15633–e15633.
-
- Chu CRS, Pietzak E. Immune mechanisms and molecular therapeutic strategies to enhance immunotherapy in non-muscle invasive bladder cancer Invited review for special issue "Seminar: Treatment Advances and Molecular Biology Insights in Urothelial Carcinoma". Urol Oncol-Semin Ori. 2023;41:398–409. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

