The modulation of calcium and chloride channels induces cardiomyocytes from human pluripotent stem cells
- PMID: 39744425
- PMCID: PMC11667823
- DOI: 10.7150/ijbs.95568
The modulation of calcium and chloride channels induces cardiomyocytes from human pluripotent stem cells
Abstract
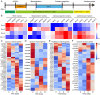
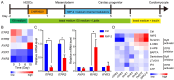
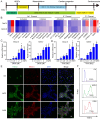
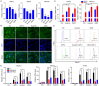
Ion channels play a crucial role in cardiac functions, and their activities exhibit dynamic changes during heart development. However, the precise function of ion channels in human heart development remains elusive. In this study, we utilized human embryonic stem cells (hESCs) as a model to mimic the process of human embryonic heart development. During hESCs differentiation into cardiomyocytes, we observed differential expression of ion channel genes, including upregulation of ryanodine receptor 2 (RYR2), which encodes a calcium release channel. Subsequently, we discovered that Suramin, an activator of RyR2, efficiently promoted cardiac differentiation even in the absence of conventional WNT inhibitors. Furthermore, various modulators targeting sodium channels, potassium channels or chloride channels were examined under chemically defined conditions during cardiac differentiation. We found that DIDS, a chloride transport inhibitor, also enhanced hESCs differentiation into cardiomyocytes. Both Suramin and DIDS partially inhibited WNT signaling pathway, and RYR2 knockdown attenuated cardiac differentiation induced by WNT inhibitor treatment, or Suramin or DIDS administration. The resulting cardiomyocytes induced by these ion modulators exhibited specific expression patterns of cardiac genes and displayed typical electrophysiological signals. Notably, compared to WNT inhibitor treatment group, both Suramin and DIDS led to increased generation of atrial-like cardiomyocytes suggesting their potential as alternative inducers for specific cardiomyocyte lineage commitment during human cardiomyocyte induction processes. This study demonstrates that regulation of ion channels plays a crucial role in determining the fate of cardiac cells, providing an effective approach for inducing cardiomyocytes from hPSCs and highlighting their critical involvement in human heart development.
Keywords: calcium channel; cardiomyocyte derivation; chloride channel; human pluripotent stem cells; ryanodine receptor 2.
© The author(s).
Conflict of interest statement
Competing Interests: YM and GC filed a patent application based on cardiomyocyte differentiation that is induced by chloride channel regulators. HELP Stem Cell Innovations Co. Ltd. performed the electrophysiological analysis of hESC-derived cardiomyocytes.
Figures


Similar articles
-
Similarities in the effects of DIDS, DBDS and suramin on cardiac ryanodine receptor function.J Membr Biol. 1999 Mar 15;168(2):159-68. doi: 10.1007/s002329900506. J Membr Biol. 1999. PMID: 10089236
-
Volume-sensitive outwardly rectifying chloride channel blockers protect against high glucose-induced apoptosis of cardiomyocytes via autophagy activation.Sci Rep. 2017 Mar 16;7:44265. doi: 10.1038/srep44265. Sci Rep. 2017. PMID: 28300155 Free PMC article.
-
RyR2 modulates a Ca2+-activated K+ current in mouse cardiac myocytes.PLoS One. 2014 Apr 18;9(4):e94905. doi: 10.1371/journal.pone.0094905. eCollection 2014. PLoS One. 2014. PMID: 24747296 Free PMC article.
-
Calcium signaling in human stem cell-derived cardiomyocytes: Evidence from normal subjects and CPVT afflicted patients.Cell Calcium. 2016 Mar;59(2-3):98-107. doi: 10.1016/j.ceca.2015.12.002. Epub 2015 Dec 15. Cell Calcium. 2016. PMID: 26725479 Free PMC article. Review.
-
Lessons from the heart: mirroring electrophysiological characteristics during cardiac development to in vitro differentiation of stem cell derived cardiomyocytes.J Mol Cell Cardiol. 2014 Feb;67:12-25. doi: 10.1016/j.yjmcc.2013.12.011. Epub 2013 Dec 23. J Mol Cell Cardiol. 2014. PMID: 24370890 Review.
References
-
- Davies MP, An RH, Doevendans P, Kubalak S, Chien KR, Kass RS. Developmental changes in ionic channel activity in the embryonic murine heart. Circ Res. 1996;78:15–25. - PubMed
-
- Conforti L, Tohse N, Sperelakis N. Tetrodotoxin-sensitive sodium current in rat fetal ventricular myocytes-contribution to the plateau phase of action potential. J Mol Cell Cardiol. 1993;25:159–73. - PubMed
-
- Boczek NJ, Ye D, Jin F, Tester DJ, Huseby A, Bos JM. et al. Identification and Functional Characterization of a Novel CACNA1C-Mediated Cardiac Disorder Characterized by Prolonged QT Intervals With Hypertrophic Cardiomyopathy, Congenital Heart Defects, and Sudden Cardiac Death. Circ Arrhythm Electrophysiol. 2015;8:1122–32. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

