Gene expression profiling in PBMCs for acute rejection in lung transplant recipients reveals myeloid responses
- PMID: 39744543
- PMCID: PMC11688322
- DOI: 10.3389/frtra.2024.1508419
Gene expression profiling in PBMCs for acute rejection in lung transplant recipients reveals myeloid responses
Abstract
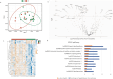
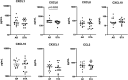
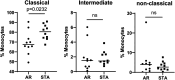
The acute rejection (AR) diagnosis depends on transbronchial biopsy, which is semi-invasive and not easily performed. Our study used the Nanostring gene expression technology on PBMCs obtained from lung transplant recipients (LTRs) to search for biomarkers. We identified distinct differential gene profiles between patients with stable status (STA) and AR. Subsequently, we independently evaluated monocyte compositions in PBMCs using flow cytometry and assessed the levels of 7 chemokines in serum using Luminex. The 48 top differentially expressed genes (DEGs) were identified, utilizing a criterion of at least a 1.5-fold change between two groups, with a false discovery rate (FDR) p-Adj < 0.05. Of these 48 genes, the top 10 genes with the highest fold changes and significant p-values were selected for qPCR validation. CD68, ANXA1, ITGB, and IFI30 can be confirmed among the validated genes. A significantly lower percentage of CD14 + CD16- classical monocytes was observed in AR than in STA patients, which aligns with downregulated DEGs. Many of the DEGs were related to monocytes-macrophages and chemokines. Although these results still need to be confirmed in larger cohorts, they suggest that gene profiling of PBMC can help to identify markers related to AR in LTRs.
Keywords: NanoString; acute rejection; gene expression; lung transplantation; monocytes.
© 2024 Liu, Westra, Hu, Verschuuren, van Kempen, van Baarle and Bos.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Mrad A, Chakraborty RK. Lung Transplant Rejection. Treasure Island (FL): StatPearls; (2021). - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

