WTAP regulates Mitochondrial damage and Lipid oxidation in HCC by NOA1 mediated m6A modification
- PMID: 39744575
- PMCID: PMC11660133
- DOI: 10.7150/jca.102618
WTAP regulates Mitochondrial damage and Lipid oxidation in HCC by NOA1 mediated m6A modification
Abstract
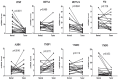
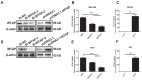
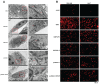
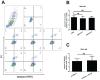
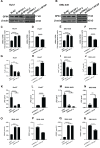
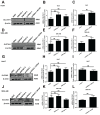
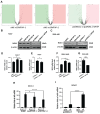
Background: Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death worldwide. However, the molecular mechanism underlying the occurrence and development of HCC remains unclear. We are interested in the function of m6A methylation enzyme WTAP in the occurrence and development of HCC. Methods: Expression of the m6A methylation-associated enzymes in paired carcinoma and adjacent tissues (N=17) were detected by RT-PCR. Electron microscopy was adopted to observe the subcellular organelle. GPX4 levels in hepatoma cells were analyzed by Western blot and RT-PCR. The Fe2+ and GSH/GSSG levels were detected using the corresponding kits. Mass spectrometry (MS) was conducted to determine the altered protein types in hepatoma cells. Finally, methylated RNA immunoprecipitation (MeRIP) was used to analyze the m6A methylation of Nitric oxide-associated protein 1 (NOA1). Results: RT-PCR showed that there were no significant differences among tumor tissues and normal tissues in METTL3 (p=0.6485), FTO (p=0.1158), ALKBH (p=0.6148), YTH N6-Methyladenosine RNA binding protein F1 (YTHDF1) (p=0.3171), and YTH N6-Methyladenosine RNA binding protein F2 (YTHDF2) (p=0.1116). However, compared to normal tissue, WTAP (p=0.0011), METLL14 (p=0.0044) and YTH N6-Methyladenosine RNA binding protein F3 (YTHDF3) (p=0.0472) were obviously decreased in tumor tissues. The decrease of WTAP was most apparent. Conditional knockout of WTAP in Huh-7 and SNU-449 cells could induce mitochondria damage, which was manifested in smaller mitochondria and a compressed intermembrane space of mitochondria. The result was also confirmed by electron microscopy. Additionally, Huh-7 and SNU-449 cells with WTAP knockdown presented low mitochondrial membrane potential, while WTAP overexpression could reverse this effect. Interestingly, data from flow cytometry by Annexin V-FITC/PI and detection of pyroptosis-related marker Gasdermin D (GSDMD) by Western blot demonstrated that, overexpressing or knocking down WTAP will not affect cell apoptosis and pyroptosis in hepatoma cells. Furthermore, mRNA and protein levels of the key indicator GPX4 of ferroptosis in Huh-7 and SNU-449 cells with WTAP knockdown or overexpression were analyzed by RT-PCR and Western blot. It was shown that knockdown of WTAP promoted expressions of GPX4 in these cells (p<0.0001), but a distinct downregulation of GPX4 levels occurred in the WTAP overexpressing cells. Further study indicated that a significantly increase of GSH/GSSG levels and clearly decrease of Fe2+ concentrations appeared in Huh-7 and SNU-449 cells with WTAP knockdown (p<0.05). Opposite results were observed in the cells with WTAP overexpression (p<0.05). Moreover, we also clarified the effect of WTAP on modulating GSH synthesis might be independent of SLC7A11, not SLC3A2 or the Xc-system. Finally, mass spectrometry results showed that NOA1 might be related to WTAP. qPCR, WB and MeRIP-qPCR also confirmed WTAP regulated the m6A methylation of NOA1. It is supposed that NOA1 might be the molecule at the heart of the regulation mechanism by WTAP. Conclusion: WTAP may affect the m6A methylation of NOA1 to induce mitochondrial damage, meanwhile activate the GPX4-axis to inhibit the lipid oxidation, resulting in the development of HCC.
Keywords: HCC; Lipid oxidation; Mitochondrial damage; NOA1; WTAP.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
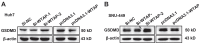
Similar articles
-
WTAP-mediated N6-methyladenosine modification promotes the inflammation, mitochondrial damage and ferroptosis of kidney tubular epithelial cells in acute kidney injury by regulating LMNB1 expression and activating NF-κB and JAK2/STAT3 pathways.J Bioenerg Biomembr. 2024 Jun;56(3):285-296. doi: 10.1007/s10863-024-10015-0. Epub 2024 Mar 22. J Bioenerg Biomembr. 2024. PMID: 38517565
-
WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1.Mol Cancer. 2019 Aug 22;18(1):127. doi: 10.1186/s12943-019-1053-8. Mol Cancer. 2019. PMID: 31438961 Free PMC article.
-
WTAP-Mediated m6A Modification of TRAIL-DR4 Suppresses MH7A Cell Apoptosis.Int J Rheum Dis. 2025 Jan;28(1):e70065. doi: 10.1111/1756-185X.70065. Int J Rheum Dis. 2025. PMID: 39797510
-
WTAP-mediated m6A modification on circCMTM3 inhibits hepatocellular carcinoma ferroptosis by recruiting IGF2BP1 to increase PARK7 stability.Dig Liver Dis. 2023 Jul;55(7):967-981. doi: 10.1016/j.dld.2022.12.005. Epub 2022 Dec 30. Dig Liver Dis. 2023. PMID: 36586770
-
METTL3 regulates m6A in endometrioid epithelial ovarian cancer independently of METTl14 and WTAP.Cell Biol Int. 2020 Dec;44(12):2524-2531. doi: 10.1002/cbin.11459. Epub 2020 Sep 11. Cell Biol Int. 2020. PMID: 32869897
Cited by
-
Predictive models and WTAP targeting for idiopathic pulmonary fibrosis (IPF).Sci Rep. 2025 Apr 26;15(1):14622. doi: 10.1038/s41598-025-98490-2. Sci Rep. 2025. PMID: 40287490 Free PMC article.
-
From mitochondrial dysregulation to ferroptosis: Exploring new strategies and challenges in radioimmunotherapy (Review).Int J Oncol. 2025 Sep;67(3):76. doi: 10.3892/ijo.2025.5781. Epub 2025 Aug 8. Int J Oncol. 2025. PMID: 40776761 Free PMC article. Review.
References
-
- Llovet JM, Kelley RK, Villanueva A, Singal AG, Pikarsky E, Roayaie S. et al. Hepatocellular carcinoma. Nat Rev Dis Primers. 2021;7:6. - PubMed
-
- Sugawara Y, Hibi T. Surgical treatment of hepatocellular carcinoma. Biosci Trends. 2021;15:138–41. - PubMed
-
- Gramantieri L, Pollutri D, Gagliardi M, Giovannini C, Quarta S, Ferracin M. et al. MiR-30e-3p Influences Tumor Phenotype through MDM2/TP53 Axis and Predicts Sorafenib Resistance in Hepatocellular Carcinoma. Cancer Res. 2020;80:1720–34. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

