Advancing crop improvement through GWAS and beyond in mung bean
- PMID: 39744597
- PMCID: PMC11688477
- DOI: 10.3389/fpls.2024.1436532
Advancing crop improvement through GWAS and beyond in mung bean
Abstract
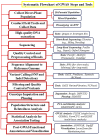
Accessing the underlying genetics of complex traits, especially in small grain pulses is an important breeding objective for crop improvement. Genome-wide association studies (GWAS) analyze thousands of genetic variants across several genomes to identify links with specific traits. This approach has discovered many strong associations between genes and traits, and the number of associated variants is expected to continue to increase as GWAS sample sizes increase. GWAS has a range of applications like understanding the genetic architecture associated with phenotype, estimating genetic correlation and heritability, developing genetic maps based on novel identified quantitative trait loci (QTLs)/genes, and developing hypotheses related to specific traits in the next generation. So far, several causative alleles have been identified using GWAS which had not been previously detected using QTL mapping. GWAS has already been successfully applied in mung bean (Vigna radiata) to identify SNPs/alleles that are used in breeding programs for enhancing yield and improvement against biotic and abiotic factors. In this review, we summarize the recently used advanced genetic tools, the concept of GWAS and its improvement in combination with structural variants, the significance of combining high-throughput phenotyping and genome editing with GWAS, and also highlights the genetic discoveries made with GWAS. Overall, this review explains the significance of GWAS with other advanced tools in the future, concluding with an overview of the current and future applications of GWAS with some recommendations.
Keywords: GWAS; QTLs; high-throughput phenotyping; mung bean; structural variants.
Copyright © 2024 Ahmed, Asghar, Hameed, Ghaffar and Shahid.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Ahmed S. R., Anwar Z., Shahbaz U., Skalicky M., Ijaz A., Tariq M. S., et al. . (2023). Potential role of silicon in plants against biotic and abiotic stresses. Silicon 7, 3283–3303. doi: 10.1007/s12633-022-02254-w - DOI
Publication types
LinkOut - more resources
Full Text Sources

