A single-cell compendium of human cerebrospinal fluid identifies disease-associated immune cell populations
- PMID: 39744938
- PMCID: PMC11684814
- DOI: 10.1172/JCI177793
A single-cell compendium of human cerebrospinal fluid identifies disease-associated immune cell populations
Abstract
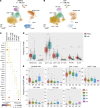
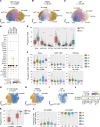
Single-cell transcriptomics applied to cerebrospinal fluid (CSF) for elucidating the pathophysiology of neurologic diseases has produced only a preliminary characterization of CSF immune cells. CSF derives from and borders central nervous system (CNS) tissue, allowing for comprehensive accounting of cell types along with their relative abundance and immunologic profiles relevant to CNS diseases. Using integration techniques applied to publicly available datasets in combination with our own studies, we generated a compendium with 139 subjects encompassing 135 CSF and 58 blood samples. Healthy subjects and individuals across a wide range of diseases, such as multiple sclerosis (MS), Alzheimer's disease, Parkinson's disease, COVID-19, and autoimmune encephalitis, were included. We found differences in lymphocyte and myeloid subset frequencies across different diseases as well as in their distribution between blood and CSF. We identified what we believe to be a new subset of AREG+ dendritic cells exclusive to the CSF that was more abundant in subjects with MS compared with healthy controls. Finally, transcriptional cell states in CSF microglia-like cells and lymphoid subsets were elucidated. Altogether, we have created a reference compendium for single-cell transcriptional profiling encompassing CSF immune cells useful to the scientific community for future studies on neurologic diseases.
Keywords: Adaptive immunity; Immunology; Innate immunity; Neurological disorders; Neuroscience.
Figures