MHC-related protein 1-restricted recognition of cancer via a semi-invariant TCR-α chain
- PMID: 39744940
- PMCID: PMC11684821
- DOI: 10.1172/JCI181895
MHC-related protein 1-restricted recognition of cancer via a semi-invariant TCR-α chain
Abstract
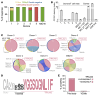
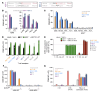
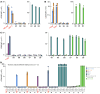
The T cell antigen presentation platform MR1 consists of 6 allomorphs in humans that differ by no more than 5 amino acids. The principal function of this highly conserved molecule involves presenting microbial metabolites to the abundant mucosal-associated invariant T (MAIT) cell subset. Recent developments suggest that the role of MR1 extends to presenting antigens from cancer cells, a function dependent on the K43 residue in the MR1 antigen binding cleft. Here, we successfully cultured cancer-activated, MR1-restricted T cells from multiple donors and confirmed that they recognized a wide range of cancer types expressing the most common MR1*01 and/or MR1*02 allomorphs (over 95% of the population), while remaining inert to healthy cells including healthy B cells and monocytes. Curiously, in all but one donor these T cells were found to incorporate a conserved TCR-α chain motif, CAXYGGSQGNLIF (where X represents 3-5 amino acids), because of pairing between 10 different TRAV genes and the TRAJ42 gene segment. This semi-invariance in the TCR-α chain is reminiscent of MAIT cells and suggests recognition of a conserved antigen bound to K43.
Keywords: Cancer; Cellular immune response; Immunology; Oncology; T cells.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

