Whole-exome sequencing association study reveals genetic effects on tumor microenvironment components in nasopharyngeal carcinoma
- PMID: 39744943
- PMCID: PMC11684818
- DOI: 10.1172/JCI182768
Whole-exome sequencing association study reveals genetic effects on tumor microenvironment components in nasopharyngeal carcinoma
Abstract
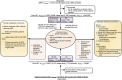
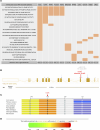
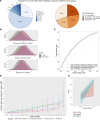
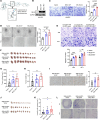
Nasopharyngeal carcinoma (NPC) presents a substantial clinical challenge due to the limited understanding of its genetic underpinnings. Here we conduct the largest scale whole-exome sequencing association study of NPC to date, encompassing 6,969 NPC cases and 7,100 controls. We unveil 3 germline genetic variants linked to NPC susceptibility: a common rs2276868 in RPL14, a rare rs5361 in SELE, and a common rs1050462 in HLA-B. We also underscore the critical impact of rare genetic variants on NPC heritability and introduce a refined composite polygenic risk score (rcPRS), which outperforms existing models in predicting NPC risk. Importantly, we reveal that the polygenic risk for NPC is mediated by EBV infection status. Utilizing a comprehensive multiomics approach that integrates both bulk-transcriptomic (n = 356) and single-cell RNA sequencing (n = 56) data with experimental validations, we demonstrate that the RPL14 variant modulates the EBV life cycle and NPC pathogenesis. Furthermore, our data indicate that the SELE variant contributes to modifying endothelial cell function, thereby facilitating NPC progression. Collectively, our study provides crucial insights into the intricate genetic architecture of NPC, spotlighting the vital interplay between genetic variations and tumor microenvironment components, including EBV and endothelial cells, in predisposing to NPC. This study opens new avenues for advancements in personalized risk assessments, early diagnosis, and targeted therapies for NPC.
Keywords: Cancer; Genetic variation; Genetics; Oncology.
Figures




References
-
- Chen C, et al. Multiple risk factors of nasopharyngeal carcinoma: Epstein-Barr virus, malarial infection, cigarette smoking and familial tendency. Anticancer Res. 1990;10(2b):547–553. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

