Highly Pathogenic Avian Influenza (HPAI) H5N1 virus in Finland in 2021-2023 - Genetic diversity of the viruses and infection kinetics in human dendritic cells
- PMID: 39745171
- PMCID: PMC11727053
- DOI: 10.1080/22221751.2024.2447618
Highly Pathogenic Avian Influenza (HPAI) H5N1 virus in Finland in 2021-2023 - Genetic diversity of the viruses and infection kinetics in human dendritic cells
Abstract
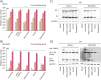
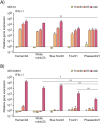
Highly pathogenic avian influenza (HPAI) H5N1 is known for its virulence and zoonotic potential, infecting birds and mammals, thus raising public health concerns. Since 2021 its spread among birds has led to cross-species transmission causing epizootics among mammals, eventually impacting fur animal farms in Finland in 2023. To analyze the infectivity of the Finnish H5N1 isolates in human cells, representatives of diverse H5N1 isolates were selected based on the genetic differences, host animal species, and the year of occurrence. The infection kinetics of the selected H5N1 isolates from wild pheasant and fox, and fur animals blue fox and white mink were examined in human monocyte-derived dendritic cells (moDCs) with H5N1 human isolate as a control. Although the isolate from pheasant (a wild bird) showed weakly reduced replication and viral protein expression in human cells compared to mammalian isolates, no discernible differences in virus replication in moDCs was observed. This study revealed similar infectivity in human moDCs for all five H5N1 isolates, regardless of the observed genetic differences. While H5N1 human infections remain rare, the virus poses a risk for widespread epizootics in mammals such as fur animal farms and, more recently, dairy cattle.
Keywords: Finland; H5N1; genetic variation; highly pathogenic avian influenza (HPAI); human cells; immune cells; infectivity; viral kinetic.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
-
- CDC . Avian Influenza (Bird Flu). 2024 [cited 2024 Nov 13]. Past reported global human cases with highly pathogenic avian influenza A (H5N1) (HPAI H5N1) by country, 1997–2024. Available from: https://www.cdc.gov/bird-flu/php/avian-flu-summary/chart-epi-curve-ah5n1....
-
- Tammiranta N, Isomursu M, Fusaro A, et al. . Highly pathogenic avian influenza A (H5N1) virus infections in wild carnivores connected to mass mortalities of pheasants in Finland. Infect Genet Evol. 2023 Jul 1;111:105423. - PubMed
-
- Finnish Food Authority [Internet]. [cited 2024 Nov 13]. Avian influenza in Finland. Available from: https://www.ruokavirasto.fi/en/animals/animal-health-and-diseases/animal....
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
