Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii group in Taiwan
- PMID: 39745372
- PMCID: PMC11774041
- DOI: 10.1128/msphere.00793-24
Molecular epidemiology of carbapenem-resistant Acinetobacter baumannii group in Taiwan
Abstract
Acinetobacter, particularly the Acinetobacter baumannii group, is a major cause of nosocomial infections, and carbapenem-resistant Acinetobacter spp. are important human pathogens. We collected 492 Acinetobacter spp. strains from two hospitals in Taiwan and classified them using MALDI-TOF MS and blaOXA-51-like PCR; 94.5% were A. baumannii, and 5.5% were non-A. baumannii (NAB). We confirmed their identity by rpoB gene sequencing of 239 randomly selected A. baumannii strains and all 27 NAB strains. Our analysis revealed that the rpoB alleles of OXA51-like-negative strains matched those of two NAB species, A. seifertii and A. nosocomialis, while all OXA51-like-positive strains matched A. baumannii, as per the Pasteur MLST scheme database. Among the 492 strains, 240 exhibited carbapenem resistance, including 237 carbapenem-resistant A. baumannii (CRAB) strains and three CR-NAB strains. All CRAB strains were positive for blaOXA-51-like; 72.6% also carried blaOXA-23-like, 22.8% carried blaOXA-24-like, 3.4% co-carried blaOXA-23-like+blaOXA-24-like, and 1.27% carried blaOXA-51-like alone. Among the three CR-NAB strains, one carried blaNDM-1, and two co-carried blaOXA-58-like+blaIMP. We also established a new multiplex PCR method for rapid screening of common capsular types (KL), which showed a difference between CRAB and carbapenem-susceptible A. baumannii (CSAB). KL2, KL10, KL22, and KL52 accounted for 76.6% of CRAB strains, whereas about half of the CSAB strains were other KL types. Of the remaining CSAB strains, KL14 was the most predominant type at 10.3%. We further conducted MLST Pasteur typing for 262 isolates and found that the carbapenemase genes were correlated with either ST or KL types. Additionally, KL types showed correlations with ST types, carbapenem resistance, and certain clinical records. Whole-genome sequencing of a blaNDM-1-carrying A. seifertii strain revealed a plasmid transferable via in vitro conjugation, suggesting A. seifertii may be a reservoir for NDM-1 plasmids.IMPORTANCECarbapenem-resistant Acinetobacter spp. have been identified by the World Health Organization as a top priority for new antibiotic development. We established a rapid KL-typing method for efficient screening of Acinetobacter baumannii strains to enable epidemiological surveillance and provide a foundation for effective infection control. Our investigation of the molecular epidemiology of the A. baumannii group isolates revealed the prevalence of carbapenemase genes and major KL types among CR and CS strains of A. baumannii and NAB. We identified an A. seifertii strain carrying a Ti-type conjugative operon on a small plasmid that harbored genes encoding the NDM-1 carbapenemase alongside genes conferring resistance to aminoglycosides and bleomycin and closely resembled sequences detected in A. soli and A. pittii in Taiwan and China, respectively, suggesting its potential for transmitting multidrug resistance and contributing to the spread of antimicrobial resistance.
Keywords: Acinetobacter baumannii group; capsular types; carbapenemase genes; wzy multiplex PCR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

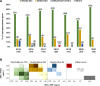

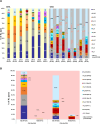

References
-
- Cosgaya C, Marí-Almirall M, Van Assche A, Fernández-Orth D, Mosqueda N, Telli M, Huys G, Higgins PG, Seifert H, Lievens B, Roca I, Vila J. 2016. Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int J Syst Evol Microbiol 66:4105–4111. doi: 10.1099/ijsem.0.001318 - DOI - PubMed
-
- Nemec A, Krizova L, Maixnerova M, van der Reijden TJK, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006 - DOI - PubMed
-
- Esterly JS, Griffith M, Qi C, Malczynski M, Postelnick MJ, Scheetz MH. 2011. Impact of carbapenem resistance and receipt of active antimicrobial therapy on clinical outcomes of Acinetobacter baumannii bloodstream infections. Antimicrob Agents Chemother 55:4844–4849. doi: 10.1128/AAC.01728-10 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- MOST 110-2320-B-039-059/National Science and Technology Council (NSTC)
- DMR-112-145/China Medical University Hospital (CMUH)
- DMR-113-123/China Medical University Hospital (CMUH)
- CMU112-S-46/China Medical University, Taiwan (CMU)
- CMRPG3K1011/Chang Gung Memorial Hospital, Linkou (Linkou Chang Gung Memorial Hospital)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

