mRNA-LNP vaccines combined with tPA signal sequence elicit strong protective immunity against Klebsiella pneumoniae
- PMID: 39745376
- PMCID: PMC11774038
- DOI: 10.1128/msphere.00775-24
mRNA-LNP vaccines combined with tPA signal sequence elicit strong protective immunity against Klebsiella pneumoniae
Abstract
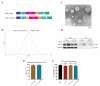
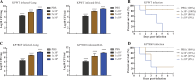
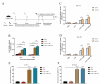
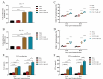
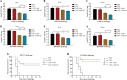
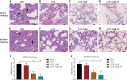
Klebsiella pneumoniae is a prominent Gram-negative and encapsulated opportunistic pathogen that causes a multitude of infections such as severe respiratory and healthcare-associated infections. Despite the widespread anti-microbial resistance and the high mortality rate, currently, no clinically vaccine is approved for battling K. pneumoniae. To date, messenger RNA (mRNA) vaccine is one of the most advancing technologies and are extensively investigated for viral infection, while infrequently applied for prevention of bacterial infections. In the present study, we aim to construct a new mRNA vaccine encoding YidR or combining with a tissue plasminogen activator signal sequence for preventing K. pneumoniae infection. Adaptive immunity was determined in mRNA vaccines-immunized mice and the protective effects of mRNA vaccines were evaluated in K. pneumoniae infected models. The results showed that lipid nanoparticle (LNP)-YidR-mRNA vaccine was produced with good morphology, high the encapsulation efficiency, and the specific antigen was highly expressed in cells in vitro. In addition, immunization with either LNP-YidR or LNP-YidR-SP elicited a Th1-biased immune response, reduced bacterial load, and provided broad protection in the lung infection models. Importantly, the LNP-YidR-SP mRNA vaccine induced strong adaptive humoral and cellular immunity and increased the survivability of mice compared to the other groups. Our findings serve as a focal point for developing a potential mRNA vaccine against K. pneumoniae, indicating the potential of mRNA vaccines for improving next-generation bacterial vaccine.IMPORTANCEK. pneumoniae is a notorious and clinical bacterium that is evolving in community-acquired and nosocomial settings. This opportunistic pathogen causes severe infectious diseases, including urinary tract infection and pneumonia, and causes a concerning global public burden. Despite efforts having been created to develop different types of K. pneumoniae vaccines, there is no licensed vaccine for preventing K. pneumoniae infection. Therefore, to develop an effective tactic is essential to combat K. pneumoniae-caused diseases. This study provides a novel vaccine strategy against K. pneumoniae and a potent platform to elicit high levels of humoral and cell-meditated immunity.
Keywords: Klebsiella pneumoniae; antibacterial immunity; mRNA vaccine; virulence.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
