Are we serologically prepared against an avian influenza pandemic and could seasonal flu vaccines help us?
- PMID: 39745389
- PMCID: PMC11796349
- DOI: 10.1128/mbio.03721-24
Are we serologically prepared against an avian influenza pandemic and could seasonal flu vaccines help us?
Abstract
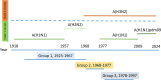
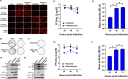
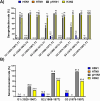
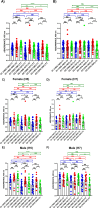
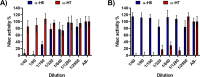
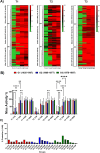
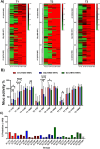
The current situation with H5N1 highly pathogenic avian influenza virus (HPAI) is causing a worldwide concern due to multiple outbreaks in wild birds, poultry, and mammals. Moreover, multiple zoonotic infections in humans have been reported. Importantly, HPAI H5N1 viruses with genetic markers of adaptation to mammals have been detected. Together with HPAI H5N1, avian influenza viruses H7N9 (high and low pathogenic) stand out due to their high mortality rates in humans. This raises the question of how prepared we are serologically and whether seasonal vaccines are capable of inducing protective immunity against these influenza subtypes. An observational study was conducted in which sera from people born between years 1925-1967, 1968-1977, and 1978-1997 were collected before or after 28 days or 6 months post-vaccination with an inactivated seasonal influenza vaccine. Then, hemagglutination inhibition, viral neutralization, and immunoassays were performed to assess the basal protective immunity of the population as well as the ability of seasonal influenza vaccines to induce protective responses. Our results indicate that subtype-specific serological protection against H5N1 and H7N9 in the representative Spanish population evaluated was limited or nonexistent. However, seasonal vaccination was able to increase the antibody titers to protective levels in a moderate percentage of people, probably due to cross-reactive responses. These findings demonstrate the importance of vaccination and suggest that seasonal influenza vaccines could be used as a first line of defense against an eventual pandemic caused by avian influenza viruses, to be followed immediately by the use of more specific pandemic vaccines.IMPORTANCEInfluenza A viruses (IAV) can infect and replicate in multiple mammalian and avian species. Avian influenza virus (AIV) is a highly contagious viral disease that occurs primarily in poultry and wild water birds. Due to the lack of population immunity in humans and ongoing evolution of AIV, there is a continuing risk that new IAV could emerge and rapidly spread worldwide, causing a pandemic, if the ability to transmit efficiently among humans was gained. The aim of this study is to analyze the basal protection and presence of antibodies against IAV H5N1 and H7N9 subtypes in the population from different ages. Moreover, we have evaluated the humoral response after immunization with a seasonal influenza vaccine. This study is strategically important to evaluate the level of population immunity that is a major factor when assessing the impact that an emerging IAV strain would have, and the role of seasonal vaccines to mitigate the effects of a pandemic.
Keywords: avian influenza; influenza A virus; influenza vaccine; nanoluciferase; neutralizing antibodies; pandemic; seroconversion; seroprotection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Evaluation of in vitro cross-reactivity to avian H5N1 and pandemic H1N1 2009 influenza following prime boost regimens of seasonal influenza vaccination in healthy human subjects: a randomised trial.PLoS One. 2013;8(3):e59674. doi: 10.1371/journal.pone.0059674. Epub 2013 Mar 26. PLoS One. 2013. PMID: 23555741 Free PMC article. Clinical Trial.
-
Antibody titer has positive predictive value for vaccine protection against challenge with natural antigenic-drift variants of H5N1 high-pathogenicity avian influenza viruses from Indonesia.J Virol. 2015 Apr;89(7):3746-62. doi: 10.1128/JVI.00025-15. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609805 Free PMC article.
-
Cross-Protection by Inactivated H5 Prepandemic Vaccine Seed Strains against Diverse Goose/Guangdong Lineage H5N1 Highly Pathogenic Avian Influenza Viruses.J Virol. 2020 Nov 23;94(24):e00720-20. doi: 10.1128/JVI.00720-20. Print 2020 Nov 23. J Virol. 2020. PMID: 32999029 Free PMC article.
-
Pandemic preparedness through vaccine development for avian influenza viruses.Hum Vaccin Immunother. 2024 Dec 31;20(1):2347019. doi: 10.1080/21645515.2024.2347019. Epub 2024 May 28. Hum Vaccin Immunother. 2024. PMID: 38807261 Free PMC article. Review.
-
[Influenza, an existing public health problem].Salud Publica Mex. 2006 May-Jun;48(3):244-67. doi: 10.1590/s0036-36342006000300009. Salud Publica Mex. 2006. PMID: 16813133 Review. Spanish.
Cited by
-
Seasonal Influenza Vaccination in People who Have Contact With Birds.Influenza Other Respir Viruses. 2025 Apr;19(4):e70101. doi: 10.1111/irv.70101. Influenza Other Respir Viruses. 2025. PMID: 40251878 Free PMC article.
-
A live attenuated NS1-deficient vaccine candidate for cattle-origin influenza A (H5N1) clade 2.3.4.4.b viruses.NPJ Vaccines. 2025 Jul 12;10(1):151. doi: 10.1038/s41541-025-01207-9. NPJ Vaccines. 2025. PMID: 40652001 Free PMC article.
-
Highly pathogenic avian influenza H5N1 in the United States: recent incursions and spillover to cattle.Npj Viruses. 2025 Jul 5;3(1):54. doi: 10.1038/s44298-025-00138-5. Npj Viruses. 2025. PMID: 40617962 Free PMC article. Review.
-
Neutralizing activity against bovine H5N1 HPAIV (clade 2.3.4.4b) in human plasma after seasonal influenza vaccination.Emerg Microbes Infect. 2025 Dec;14(1):2528539. doi: 10.1080/22221751.2025.2528539. Epub 2025 Jul 25. Emerg Microbes Infect. 2025. PMID: 40591910 Free PMC article.
-
Harnessing Antiviral Peptides: From Molecular Mechanisms to Clinical Translation.Curr Res Pharmacol Drug Discov. 2025 Jul 15;9:100228. doi: 10.1016/j.crphar.2025.100228. eCollection 2025. Curr Res Pharmacol Drug Discov. 2025. PMID: 40718095 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

