Deciphering the biosynthetic landscape of biofilms in glacier-fed streams
- PMID: 39745394
- PMCID: PMC11834409
- DOI: 10.1128/msystems.01137-24
Deciphering the biosynthetic landscape of biofilms in glacier-fed streams
Abstract
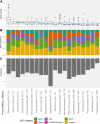
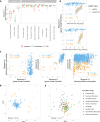
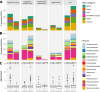
Glacier-fed streams are permanently cold, ultra-oligotrophic, and physically unstable environments, yet microbial life thrives in benthic biofilm communities. Within biofilms, microorganisms rely on secondary metabolites for communication and competition. However, the diversity and genetic potential of secondary metabolites in glacier-fed stream biofilms remain poorly understood. In this study, we present the first large-scale exploration of biosynthetic gene clusters (BGCs) from benthic glacier-fed stream biofilms sampled by the Vanishing Glaciers project from the world's major mountain ranges. We found a remarkable diversity of BGCs, with more than 8,000 of them identified within 2,868 prokaryotic metagenome-assembled genomes, some of them potentially conferring ecological advantages, such as UV protection and quorum sensing. The BGCs were distinct from those sourced from other aquatic microbiomes, with over 40% of them being novel. The glacier-fed stream BGCs exhibited the highest similarity to BGCs from glacier microbiomes. BGC composition displayed geographic patterns and correlated with prokaryotic alpha diversity. We also found that BGC diversity was positively associated with benthic chlorophyll a and prokaryotic diversity, indicative of more biotic interactions in more extensive biofilms. Our study provides new insights into a hitherto poorly explored microbial ecosystem, which is now changing at a rapid pace as glaciers are shrinking due to climate change.
Importance: Glacier-fed streams are characterized by low temperatures, high turbidity, and high flow. They host a unique microbiome within biofilms, which form the foundation of the food web and contribute significantly to biogeochemical cycles. Our investigation into secondary metabolites, which likely play an important role in these complex ecosystems, found a unique genetic potential distinct from other aquatic environments. We found the potential to synthesize several secondary metabolites, which may confer ecological advantages, such as UV protection and quorum sensing. This biosynthetic diversity was positively associated with the abundance and complexity of the microbial community, as well as concentrations of chlorophyll a. In the face of climate change, our study offers new insights into a vanishing ecosystem.
Keywords: biofilms; glacier-fed streams; microbiomes; secondary metabolites.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Fodelianakis S, Washburne AD, Bourquin M, Pramateftaki P, Kohler TJ, Styllas M, Tolosano M, De Staercke V, Schön M, Busi SB, Brandani J, Wilmes P, Peter H, Battin TJ. 2022. Microdiversity characterizes prevalent phylogenetic clades in the glacier-fed stream microbiome. ISME J 16:666–675. doi: 10.1038/s41396-021-01106-6 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

