Increasing rates of erm(B) and erm(N) in human Campylobacter coli and Campylobacter jejuni erythromycin-resistant isolates between 2018 and 2023 in France
- PMID: 39745413
- PMCID: PMC11823653
- DOI: 10.1128/aac.01668-24
Increasing rates of erm(B) and erm(N) in human Campylobacter coli and Campylobacter jejuni erythromycin-resistant isolates between 2018 and 2023 in France
Abstract
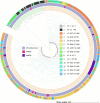
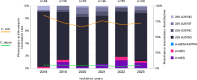
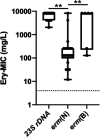
Macrolides are the first-line compounds used for the treatment of campylobacteriosis. Macrolide resistance remains low in France, with mutations in 23S rDNA being the main associated resistance mechanism. However, two erythromycin methyltransferases have also been identified: erm(B), which is mainly described in animal reservoirs, and erm(N), which is strictly described in humans. In France, between 2018 and 2023, erythromycin-resistant Campylobacter species strains were systematically sequenced and analyzed via an in-house bioinformatics pipeline, leading to the identification of the resistomes, MLST and cgMLST, as well as the characterization of the source of contamination. In this study, the genomes of 280 erythromycin-resistant strains were sequenced over a 6-year period. The identification of erythromycin-associated resistance markers revealed a predominance of 23S rDNA mutations, in 90% of cases, but also erm-type methyltransferases in 10% of cases: 75% for erm(N) and 25% for erm(B). Over this period, an important increase in the rate of erm-positive isolates was observed: 2% in 2018 compared with 13% in 2023, with 10% for erm(N) and 3% for erm(B). erm(N) has been found exclusively within a CRISPR-Cas9 operon, whereas erm(B) has been found within diverse types of resistance genomic islands. Each erm(N)- or erm(B)-positive isolate had at least two other resistance markers (mostly ciprofloxacin, tetracycline, or ampicillin) and often carried aminoglycoside-associated resistance genes. The majority of the erm-positive isolates were obtained from chicken. The increasing rates of erm-positive and multiresistant isolates make the monitoring of erythromycin-resistant Campylobacter strains, specifically within the chicken meat production, a topic of serious importance.
Keywords: Campylobacter; NGS; macrolide; methyltransferase; resistance.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Emergence of Erythromycin Resistance Methyltransferases in Campylobacter coli Strains in France.Antimicrob Agents Chemother. 2021 Oct 18;65(11):e0112421. doi: 10.1128/AAC.01124-21. Epub 2021 Aug 9. Antimicrob Agents Chemother. 2021. PMID: 34370579 Free PMC article.
-
Lack of Evidence for erm(B) Infiltration Into Erythromycin-Resistant Campylobacter coli and Campylobacter jejuni from Commercial Turkey Production in Eastern North Carolina: A Major Turkey-Growing Region in the United States.Foodborne Pathog Dis. 2018 Nov;15(11):698-700. doi: 10.1089/fpd.2018.2477. Epub 2018 Aug 10. Foodborne Pathog Dis. 2018. PMID: 30096008
-
Macrolide resistance in Campylobacter jejuni and Campylobacter coli: molecular mechanism and stability of the resistance phenotype.Antimicrob Agents Chemother. 2005 Jul;49(7):2753-9. doi: 10.1128/AAC.49.7.2753-2759.2005. Antimicrob Agents Chemother. 2005. PMID: 15980346 Free PMC article.
-
The Current State of Macrolide Resistance in Campylobacter spp.: Trends and Impacts of Resistance Mechanisms.Appl Environ Microbiol. 2017 May 31;83(12):e00416-17. doi: 10.1128/AEM.00416-17. Print 2017 Jun 15. Appl Environ Microbiol. 2017. PMID: 28411226 Free PMC article. Review.
-
Macrolide resistance in Campylobacter jejuni and Campylobacter coli.J Antimicrob Chemother. 2006 Aug;58(2):243-55. doi: 10.1093/jac/dkl210. Epub 2006 May 30. J Antimicrob Chemother. 2006. PMID: 16735431 Review.
Cited by
-
Genome Analysis of the Multidrug-Resistant Campylobacter coli BCT3 of the Sequence Type (ST) 872 Isolated from a Pediatric Diarrhea Case.Microorganisms. 2025 Jun 18;13(6):1420. doi: 10.3390/microorganisms13061420. Microorganisms. 2025. PMID: 40572309 Free PMC article.
References
-
- The European Union One Health 2022 Zoonoses Report. 2023. EFSA Journal - Wiley Online Library. Available from: https://efsa.onlinelibrary.wiley.com/doi/full/10.2903/j.efsa.2023.8442. Retrieved Dec 9 Dec 2024. - DOI
-
- Van CD. 2018. Article - Bulletin épidémiologique hebdomadaire. Available from: http://beh.santepubliquefrance.fr/beh/2018/1/2018_1_1.html. Retrieved 20 Aug 2024.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

